Molecular Docking with GNINA 1.0
David Ryan Koes
Royal Society of Chemistry Chemical Information & Computer Applications Group
David Ryan Koes
Royal Society of Chemistry Chemical Information & Computer Applications Group
May 27, 2021
Get Started: https://colab.research.google.com/drive/1GXmk1v8C-c4UtyKFqIm9HnsrVYH0pI-c
%%html
<style>
div.prompt {display:none}
div.output_subarea {max-width: 100%}
</style>
<script>
$3Dmolpromise = new Promise((resolve, reject) => {
require(['https://3dmol.org/build/3Dmol-nojquery.js'], function(){
resolve();});
});
require(['https://cdnjs.cloudflare.com/ajax/libs/Chart.js/2.2.2/Chart.js'], function(Ch){
Chart = Ch;
});
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
//the callback is provided a canvas object and data
var chartmaker = function(canvas, labels, data) {
var ctx = $(canvas).get(0).getContext("2d");
var dataset = {labels: labels,
datasets:[{
data: data,
backgroundColor: "rgba(150,64,150,0.5)",
fillColor: "rgba(150,64,150,0.8)",
}]};
var myBarChart = new Chart(ctx,{type:'bar',data:dataset,options:{legend: {display:false},
scales: {
yAxes: [{
ticks: {
min: 0,
}
}]}}});
};
$(".input .o:contains(html)").closest('.input').hide();
</script>
<script src="https://bits.csb.pitt.edu/asker.js/lib/asker.js"></script>
Acknowledgements¶


Andrew McNutt, Paul Francoeur, Rishal Aggarwal, Tomohide Masuda, Rocco Meli, Matthew Ragoza, Jocelyn Sunseri
%%html
<div id="whydock" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#whydock';
jQuery(divid).asker({
id: divid,
question: "Why do you most want to dock?",
answers: ['Predict pose','Virtual screening','Affinity prediction',"I don't know"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
What is molecular docking?¶
Predict the most likely conformation and pose of a ligand in a protein binding site.
- Sample conformational space
- Score poses
- Ideally score equals affinity or can be used to productively rank compounds
- Score $\ne$ Free Energy
%%html
<iframe width="560" height="315" src="https://3dmol.org/tests/docking.html" title="docking" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
<script>
$(".input .o:contains(html)").closest('.input').hide();
</script>
Inherent limitations of docking¶
Docking is intended to be high-throughput and fundamentally limiting approximations are made to achieve this.
- Receptor usually kept rigid or mostly rigid (limited side-chain flexibility)
- Ligand flexibility usually limited to torsions
- No explicit solvent model
Software Lineage¶
AutoDock Vina¶
Designed and implemented by Dr. Oleg Trott at the Scripps Research Institute.
Shared no code with AutoDock.
Focus on performance. Created new scoring function optimized for pose prediction.
Open Source Apache License
Published 2009, last update (version 1.1.2) 2011
Software Lineage¶
smina
Scoring and minimization with AutoDock Vina
We forked Vina to make it easier to use, especially for custom scoring function development and ligand minimization.
(Almost) identical behavior as Autodock Vina (just easier to use).
Apache/GPL2 Open Source License
Very stable source code. In maintence mode. Features are a subset of GNINA.
!wget https://downloads.sourceforge.net/project/smina/smina.static
--2021-05-26 22:45:24-- https://downloads.sourceforge.net/project/smina/smina.static Resolving downloads.sourceforge.net (downloads.sourceforge.net)... 216.105.38.13 Connecting to downloads.sourceforge.net (downloads.sourceforge.net)|216.105.38.13|:443... connected. HTTP request sent, awaiting response... 302 Found Location: https://versaweb.dl.sourceforge.net/project/smina/smina.static [following] --2021-05-26 22:45:24-- https://versaweb.dl.sourceforge.net/project/smina/smina.static Resolving versaweb.dl.sourceforge.net (versaweb.dl.sourceforge.net)... 162.251.232.173 Connecting to versaweb.dl.sourceforge.net (versaweb.dl.sourceforge.net)|162.251.232.173|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 9853920 (9.4M) [application/octet-stream] Saving to: ‘smina.static’ smina.static 100%[===================>] 9.40M 3.37MB/s in 2.8s 2021-05-26 22:45:28 (3.37 MB/s) - ‘smina.static’ saved [9853920/9853920]
!wget https://github.com/gnina/gnina/releases/download/v1.0.1/gnina
--2021-05-26 22:45:28-- https://github.com/gnina/gnina/releases/download/v1.0.1/gnina Resolving github.com (github.com)... 140.82.113.4 Connecting to github.com (github.com)|140.82.113.4|:443... connected. HTTP request sent, awaiting response... 302 Found Location: https://github-releases.githubusercontent.com/45548146/47de2300-8bd4-11eb-8355-430c51e07fae?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20210527%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20210527T024528Z&X-Amz-Expires=300&X-Amz-Signature=6b7e83aaead5347dbedbb339144d0b968b158ad46f25ad3a9b660244011605c7&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=45548146&response-content-disposition=attachment%3B%20filename%3Dgnina&response-content-type=application%2Foctet-stream [following] --2021-05-26 22:45:28-- https://github-releases.githubusercontent.com/45548146/47de2300-8bd4-11eb-8355-430c51e07fae?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20210527%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20210527T024528Z&X-Amz-Expires=300&X-Amz-Signature=6b7e83aaead5347dbedbb339144d0b968b158ad46f25ad3a9b660244011605c7&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=45548146&response-content-disposition=attachment%3B%20filename%3Dgnina&response-content-type=application%2Foctet-stream Resolving github-releases.githubusercontent.com (github-releases.githubusercontent.com)... 185.199.111.154, 185.199.110.154, 185.199.109.154, ... Connecting to github-releases.githubusercontent.com (github-releases.githubusercontent.com)|185.199.111.154|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 562802104 (537M) [application/octet-stream] Saving to: ‘gnina’ gnina 100%[===================>] 536.73M 31.0MB/s in 14s 2021-05-26 22:45:42 (38.8 MB/s) - ‘gnina’ saved [562802104/562802104]
!du -sh smina.static gnina
9.4M smina.static 537M gnina
I wasn't kidding about the extra dependencies!
However, if you are going to use gnina frequently you should build it from source so it uses the versions of libraries installed on your system (especially CUDA) which will result in a much smaller executable.
!du -sh /usr/local/bin/gnina
43M /usr/local/bin/gnina
Running GNINA¶
!chmod +x ./gnina #make executable
!./gnina
Missing receptor.
Correct usage:
Input:
-r [ --receptor ] arg rigid part of the receptor
--flex arg flexible side chains, if any (PDBQT)
-l [ --ligand ] arg ligand(s)
--flexres arg flexible side chains specified by comma
separated list of chain:resid
--flexdist_ligand arg Ligand to use for flexdist
--flexdist arg set all side chains within specified
distance to flexdist_ligand to flexible
--flex_limit arg Hard limit for the number of flexible
residues
--flex_max arg Retain at at most the closest flex_max
flexible residues
Search space (required):
--center_x arg X coordinate of the center
--center_y arg Y coordinate of the center
--center_z arg Z coordinate of the center
--size_x arg size in the X dimension (Angstroms)
--size_y arg size in the Y dimension (Angstroms)
--size_z arg size in the Z dimension (Angstroms)
--autobox_ligand arg Ligand to use for autobox
--autobox_add arg Amount of buffer space to add to
auto-generated box (default +4 on all six
sides)
--autobox_extend arg (=1) Expand the autobox if needed to ensure the
input conformation of the ligand being
docked can freely rotate within the box.
--no_lig no ligand; for sampling/minimizing flexible
residues
Scoring and minimization options:
--scoring arg specify alternative built-in scoring
function: ad4_scoring default dkoes_fast
dkoes_scoring dkoes_scoring_old vina vinardo
--custom_scoring arg custom scoring function file
--custom_atoms arg custom atom type parameters file
--score_only score provided ligand pose
--local_only local search only using autobox (you
probably want to use --minimize)
--minimize energy minimization
--randomize_only generate random poses, attempting to avoid
clashes
--num_mc_steps arg number of monte carlo steps to take in each
chain
--num_mc_saved arg number of top poses saved in each monte
carlo chain
--minimize_iters arg (=0) number iterations of steepest descent;
default scales with rotors and usually isn't
sufficient for convergence
--accurate_line use accurate line search
--simple_ascent use simple gradient ascent
--minimize_early_term Stop minimization before convergence
conditions are fully met.
--minimize_single_full During docking perform a single full
minimization instead of a truncated
pre-evaluate followed by a full.
--approximation arg approximation (linear, spline, or exact) to
use
--factor arg approximation factor: higher results in a
finer-grained approximation
--force_cap arg max allowed force; lower values more gently
minimize clashing structures
--user_grid arg Autodock map file for user grid data based
calculations
--user_grid_lambda arg (=-1) Scales user_grid and functional scoring
--print_terms Print all available terms with default
parameterizations
--print_atom_types Print all available atom types
Convolutional neural net (CNN) scoring:
--cnn_scoring arg (=1) Amount of CNN scoring: none, rescore
(default), refinement, all
--cnn arg built-in model to use, specify
PREFIX_ensemble to evaluate an ensemble of
models starting with PREFIX:
crossdock_default2018 crossdock_default2018_
1 crossdock_default2018_2
crossdock_default2018_3
crossdock_default2018_4 default2017 dense
dense_1 dense_2 dense_3 dense_4
general_default2018 general_default2018_1
general_default2018_2 general_default2018_3
general_default2018_4 redock_default2018
redock_default2018_1 redock_default2018_2
redock_default2018_3 redock_default2018_4
--cnn_model arg caffe cnn model file; if not specified a
default model will be used
--cnn_weights arg caffe cnn weights file (*.caffemodel); if
not specified default weights (trained on
the default model) will be used
--cnn_resolution arg (=0.5) resolution of grids, don't change unless you
really know what you are doing
--cnn_rotation arg (=0) evaluate multiple rotations of pose (max 24)
--cnn_update_min_frame During minimization, recenter coordinate
frame as ligand moves
--cnn_freeze_receptor Don't move the receptor with respect to a
fixed coordinate system
--cnn_mix_emp_force Merge CNN and empirical minus forces
--cnn_mix_emp_energy Merge CNN and empirical energy
--cnn_empirical_weight arg (=1) Weight for scaling and merging empirical
force and energy
--cnn_outputdx Dump .dx files of atom grid gradient.
--cnn_outputxyz Dump .xyz files of atom gradient.
--cnn_xyzprefix arg (=gradient) Prefix for atom gradient .xyz files
--cnn_center_x arg X coordinate of the CNN center
--cnn_center_y arg Y coordinate of the CNN center
--cnn_center_z arg Z coordinate of the CNN center
--cnn_verbose Enable verbose output for CNN debugging
Output:
-o [ --out ] arg output file name, format taken from file
extension
--out_flex arg output file for flexible receptor residues
--log arg optionally, write log file
--atom_terms arg optionally write per-atom interaction term
values
--atom_term_data embedded per-atom interaction terms in
output sd data
--pose_sort_order arg (=0) How to sort docking results: CNNscore
(default), CNNaffinity, Energy
Misc (optional):
--cpu arg the number of CPUs to use (the default is to
try to detect the number of CPUs or, failing
that, use 1)
--seed arg explicit random seed
--exhaustiveness arg (=8) exhaustiveness of the global search (roughly
proportional to time)
--num_modes arg (=9) maximum number of binding modes to generate
--min_rmsd_filter arg (=1) rmsd value used to filter final poses to
remove redundancy
-q [ --quiet ] Suppress output messages
--addH arg automatically add hydrogens in ligands (on
by default)
--stripH arg remove hydrogens from molecule _after_
performing atom typing for efficiency (on by
default)
--device arg (=0) GPU device to use
--no_gpu Disable GPU acceleration, even if available.
Configuration file (optional):
--config arg the above options can be put here
Information (optional):
--help display usage summary
--help_hidden display usage summary with hidden options
--version display program version
%%html
<div id="gnsucc" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#gnsucc';
jQuery(divid).asker({
id: divid,
question: "Were you able to run gnina in colab?",
answers: ['Yes','No','Eh'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
How does it work?¶
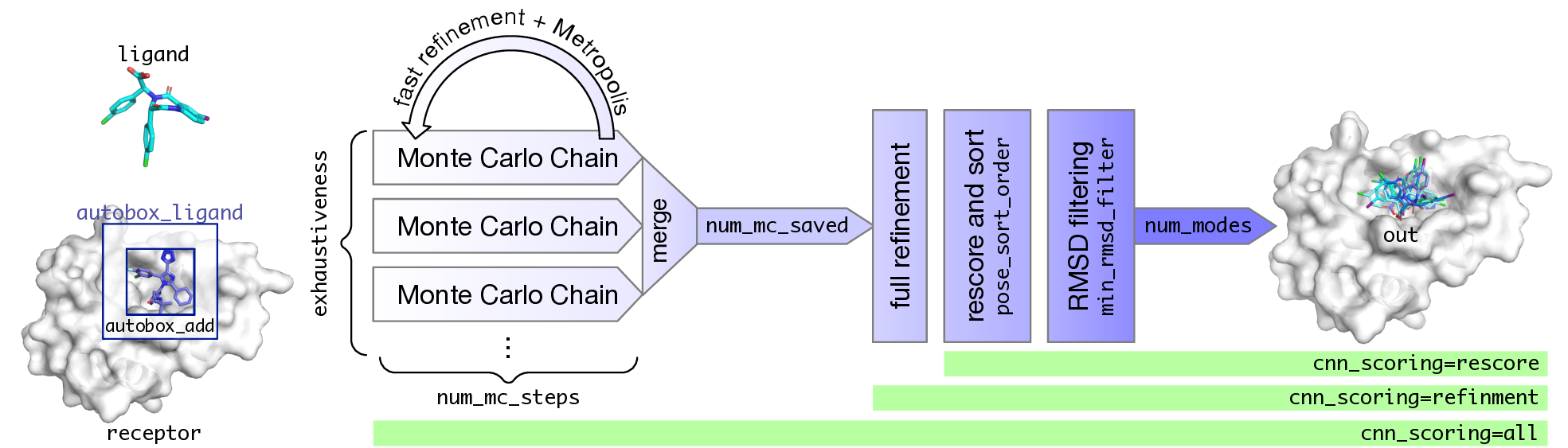
Setup Example¶
!wget http://files.rcsb.org/download/3ERK.pdb
--2021-05-26 22:45:44-- http://files.rcsb.org/download/3ERK.pdb Resolving files.rcsb.org (files.rcsb.org)... 128.6.158.70 Connecting to files.rcsb.org (files.rcsb.org)|128.6.158.70|:80... connected. HTTP request sent, awaiting response... 200 OK Length: unspecified [application/octet-stream] Saving to: ‘3ERK.pdb’ 3ERK.pdb [ <=> ] 270.37K --.-KB/s in 0.07s 2021-05-26 22:45:44 (3.68 MB/s) - ‘3ERK.pdb’ saved [276858]
!grep ATOM 3ERK.pdb > rec.pdb
!obabel rec.pdb -Orec.pdb # "sanitizing" receptor for openbabel
============================== *** Open Babel Warning in PerceiveBondOrders Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is rec.pdb) 1 molecule converted
!grep SB4 3ERK.pdb > lig.pdb
import py3Dmol
v = py3Dmol.view(height=400)
v.addModel(open('rec.pdb').read())
v.setStyle({'cartoon':{},'stick':{'radius':0.15}})
v.addModel(open('lig.pdb').read())
v.setStyle({'model':1},{'stick':{'colorscheme':'greenCarbon'}})
v.zoomTo({'model':1})
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f394c0a2d30>
Protein Preparation¶
Any file format supported by Open Babel is acceptable. Every atom in the provided file will be treated as part of the receptor.
Check for
- missing atoms
- alternative residues
- co-factors
Receptors already in a bound conformation are best, but remember to remove the ligand.
Protonation
- By default Open Babel will be used to infer protonation
- Generally only adds hydrogens
- To see what protonation will be used:
obabel rec.pdb -h -xr -Orec.pdbqt
- If PDBQT file is provided it will be taken as is with no hydrogens changed.
Ligand Preparation¶
Any file format supported by Open Babel is acceptable.
Need valid 3D conformation
Only torsions (rotatable bonds) are sampled during docking
- Ring conformations and stereoisomers are not sampled
!obabel -:'C1CNCCC1n1cnc(c2ccc(cc2)F)c1c1ccnc(n1)N' -Ol2.sdf --gen2D
1 molecule converted
This is not a valid ligand conformation (but you will still be able to dock it).
v = py3Dmol.view(height=300)
v.addModel(open('l2.sdf').read())
v.setStyle({'stick':{'colorscheme':'greenCarbon'}})
v.zoomTo()
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f394c0a28e0>
!obabel -:'C1CNCCC1n1cnc(c2ccc(cc2)F)c1c1ccnc(n1)N' -Ol3.sdf --gen3D
1 molecule converted
v = py3Dmol.view(height=400)
v.addModel(open('l3.sdf').read())
v.setStyle({'stick':{'colorscheme':'greenCarbon'}})
v.zoomTo()
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944fc17f0>
Defining the Binding Site¶
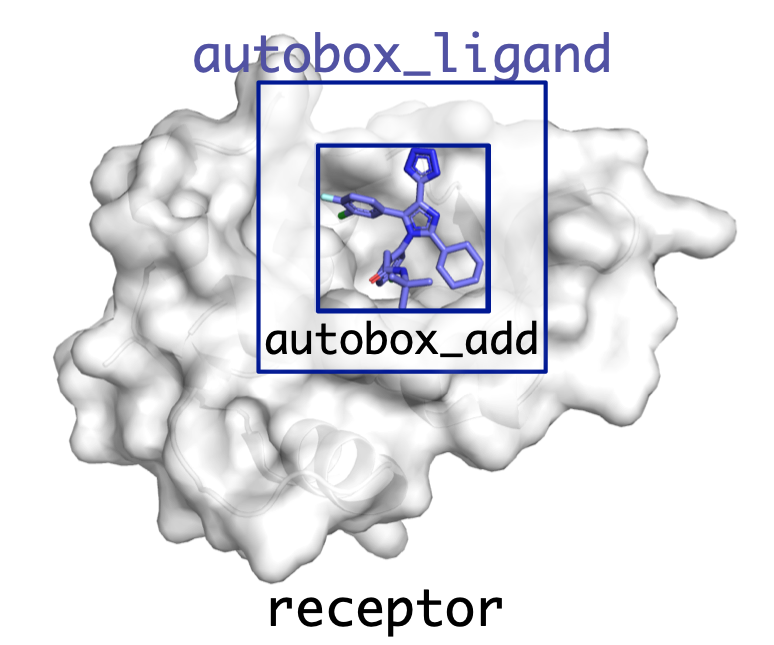
All poses are sampled within a box defined by the user.
Can be specified manually (--center_x, --size_x, etc.) but typically much easier to provide an autobox ligand.
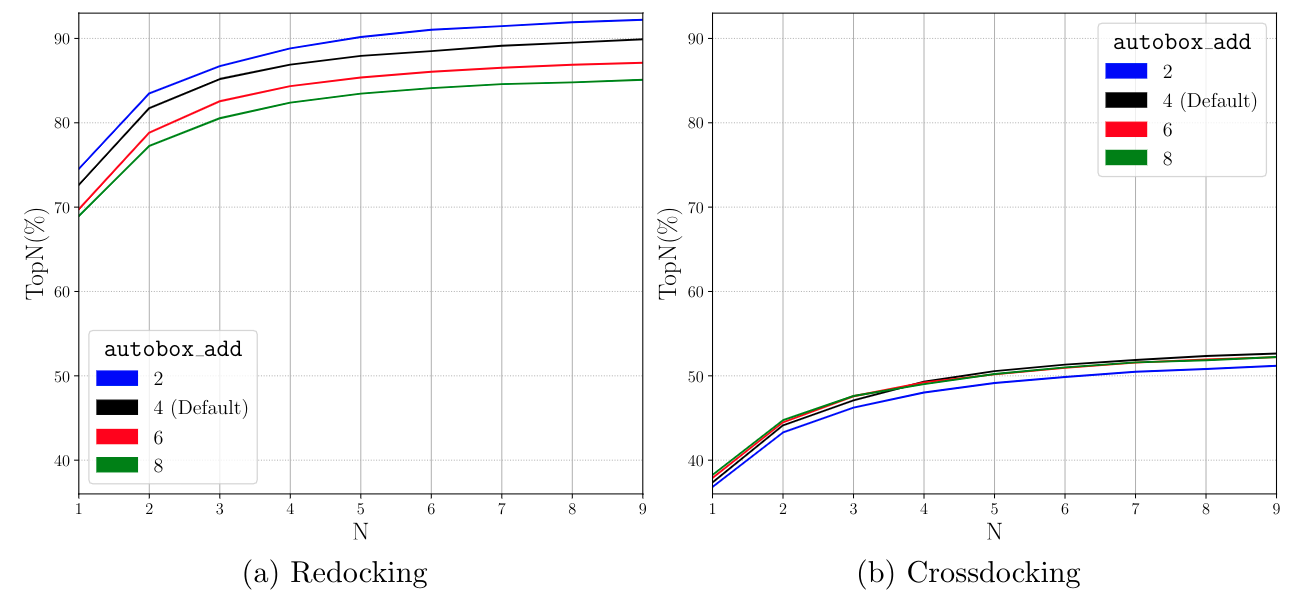
A box is created that exactly inscribes the atom coordinates of the provided ligand and then is expanded by autobox_add (default 4Å) in every dimension. If needed so provide enough room for the ligand to freely rotate the box is then further extended (autobox_extend).
Can provide any molecule for autobox_ligand (e.g. binding pocket residues, fpocket alpha spheres).
%%html
<div id="autobox" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#autobox';
jQuery(divid).asker({
id: divid,
question: "What do you think happens to docking performance when autobox_add is increased?",
answers: ['Docking is slower but better','Docking is faster and better','Docking is slower and worse','Docking is faster but worse'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
Let's Dock!¶
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb
Using random seed: -216854720
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -8.51 0.8985 6.783
2 -8.30 0.4491 6.450
3 -6.80 0.3258 6.043
4 -7.34 0.3023 6.230
5 -5.90 0.1754 5.397
6 -6.33 0.1679 5.559
7 -6.98 0.1668 5.825
8 -5.24 0.1607 5.505
9 -7.00 0.1523 5.957
Two improvements:
- Set the random seed for reproducibility (on same system)
- Specify an output file so generated poses are saved
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 -o docked.sdf.gz
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 -o docked.sdf.gz
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -8.52 0.9024 6.788
2 -8.09 0.6081 6.603
3 -8.31 0.4515 6.454
4 -6.62 0.3029 6.010
5 -6.24 0.2846 6.096
6 -6.83 0.2695 5.776
7 -6.86 0.1569 5.462
8 -6.76 0.1438 5.844
9 -6.16 0.1354 5.330
How good are the results?
We'll measure RMSD of poses with obrms from Open Babel which you can install in colab with:
!apt install openbabel
!obrms --firstonly lig.pdb docked.sdf.gz
RMSD lig.pdb: 1.44274 RMSD lig.pdb: 6.42587 RMSD lig.pdb: 6.55854 RMSD lig.pdb: 2.42936 RMSD lig.pdb: 5.64049 RMSD lig.pdb: 6.55116 RMSD lig.pdb: 4.71269 RMSD lig.pdb: 4.83259 RMSD lig.pdb: 5.52807
import gzip
v = py3Dmol.view(height=400)
v.addModel(open('rec.pdb').read())
v.setStyle({'cartoon':{},'stick':{'radius':.1}})
v.addModel(open('lig.pdb').read())
v.setStyle({'model':1},{'stick':{'colorscheme':'dimgrayCarbon','radius':.125}})
v.addModelsAsFrames(gzip.open('docked.sdf.gz','rt').read())
v.setStyle({'model':2},{'stick':{'colorscheme':'greenCarbon'}})
v.animate({'interval':1000}); v.zoomTo({'model':1}); v.rotate(90)
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944fe00a0>
%%html
<div id="betterl" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#betterl';
jQuery(divid).asker({
id: divid,
question: "If we dock a generated conformer (l3.sdf) instead, what happens to the RMSD?",
answers: ['Better','Same-ish','Worse'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
!./gnina -r rec.pdb -l l3.sdf --autobox_ligand lig.pdb --seed 0 -o docked.sdf.gz
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec.pdb -l l3.sdf --autobox_ligand lig.pdb --seed 0 -o docked.sdf.gz
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
| pose 0 | initial pose not within box
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -8.71 0.9642 6.884
2 -7.28 0.5967 6.450
3 -7.53 0.2854 6.118
4 -7.74 0.2166 6.134
5 -6.74 0.1831 5.979
6 -6.78 0.1491 5.521
7 -7.83 0.1456 5.866
8 -7.84 0.1414 5.981
9 -6.69 0.1371 5.750
!obrms --firstonly lig.pdb docked.sdf.gz
RMSD lig.pdb: 0.759809 RMSD lig.pdb: 2.13638 RMSD lig.pdb: 2.28756 RMSD lig.pdb: 4.10741 RMSD lig.pdb: 6.29767 RMSD lig.pdb: 4.78639 RMSD lig.pdb: 4.26087 RMSD lig.pdb: 4.09501 RMSD lig.pdb: 2.92727
Sampling¶
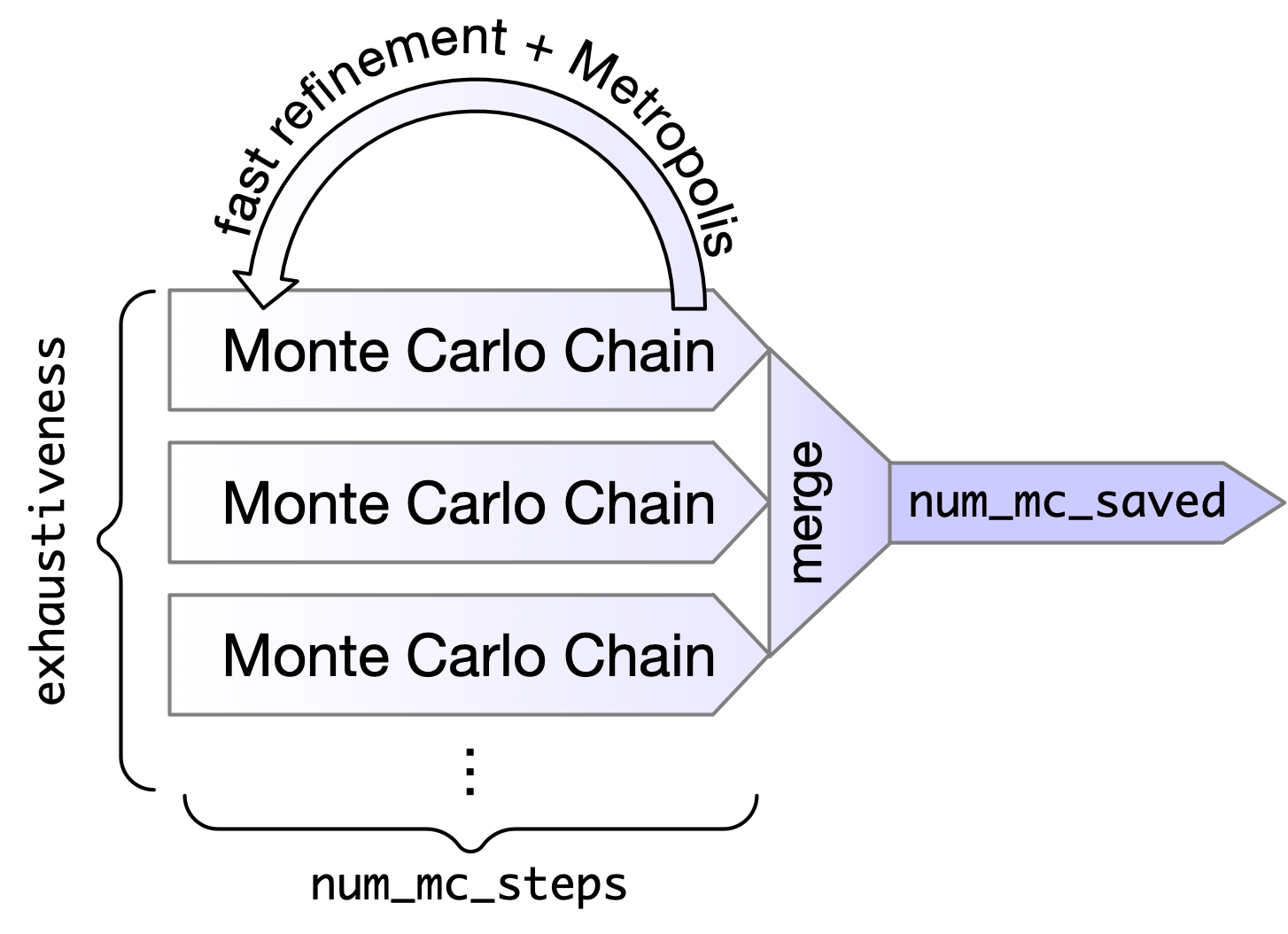
- Degrees of freedom
- 6 rigid body motions (x,y,z,pitch,yaw,roll)
- Internal torsions (not other angles/bond lengths)
- Initially randomize all degrees of freedom
- no bias to starting conformation DoF
- is biased by non-DoF conformations (e.g. ring pucker)
- Monte Carlo Chain
- Apply a random transformation (translation, rotation, or torsion)
- Perform fast refinement (truncated BFGS) of result with "soft" potentials
- Metropolis criterion to accept result as new conformation
- The more change improves conformation, more likely it is selected
- Best scoring conformations are retained.
Sampling¶

--exhaustivenessThe number of MC chains. These can be done in parallel. This is the recommended way to change the amount of sampling.--num_mc_stepsHow many iterations each MC chain performs. By default is heuristically scaled based on number of degrees of freedom (more flexible ligands will take longer). Don't recommend using unless you want to do a "quick and dirty" docking run.--num_mc_savedNumber of best scoring conformations retained by each chain and overall process. Default is max of 50 or the number of requested output conformations. Shouldn't have to change this.
Timing exhaustiveness¶
%%time
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 --exhaustiveness 1 > /dev/null 2>&1
CPU times: user 93 ms, sys: 16.8 ms, total: 110 ms Wall time: 6.7 s
MC chains are run in parallel so increasing exhaustivess won't be much slower as long as there are enough cores
%%time
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 --exhaustiveness 4 > /dev/null 2>&1
CPU times: user 83.5 ms, sys: 14.4 ms, total: 97.9 ms Wall time: 7.53 s
But if MC chains can't run in parallel expect a roughly linear increase in time.
%%time
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 --exhaustiveness 4 --cpu 1 > /dev/null 2>&1
CPU times: user 230 ms, sys: 67.3 ms, total: 297 ms Wall time: 21.5 s
For a typical docking run, there are diminishing returns in increasing the exhaustivness and the default (8) is sufficient.
Scoring¶
Empirical (e.g. Vina)
- Fast and interprettable
- A collection of weighted terms
- By default used for search and refinement
CNN
- Slower (especially without GPU) but more predictive
- By default used only for final ranking
--cnn_scoring=rescore
AutoDock Vina Scoring¶
There is no electrostatic term. Partial charges are not used. Electrostatic interactions are accounted for with hydrogen bond term.
Metals are modeled as hydrogen donors.
Terms were selected and parameterized for pose prediction performance (both speed and quality).
Final scoring function was then linearly reweighted to fit score to free energies (kcal/mol).
!./gnina --score_only -r rec.pdb -l lig.pdb --verbosity=2
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina --score_only -r rec.pdb -l lig.pdb --verbosity=2
Weights Terms
-0.035579 gauss(o=0,_w=0.5,_c=8)
-0.005156 gauss(o=3,_w=2,_c=8)
0.840245 repulsion(o=0,_c=8)
-0.035069 hydrophobic(g=0.5,_b=1.5,_c=8)
-0.587439 non_dir_h_bond(g=-0.7,_b=0,_c=8)
1.923 num_tors_div
Detected 8 CPUs
## Name gauss(o=0,_w=0.5,_c=8) gauss(o=3,_w=2,_c=8) repulsion(o=0,_c=8) hydrophobic(g=0.5,_b=1.5,_c=8) non_dir_h_bond(g=-0.7,_b=0,_c=8) num_tors_div
Reading input ... done.
Setting up the scoring function ... done.
Affinity: -8.23943 (kcal/mol)
CNNscore: 0.97413
CNNaffinity: 6.98467
CNNvariance: 0.07986
Intramolecular energy: -0.51286
Term values, before weighting:
## 84.31120 1224.00134 2.82601 33.60834 2.67216 0.00000
GPU memory usage: 1451 MB
Alternative Empirical Scoring¶
!./gnina --help | grep scoring | head -3
--scoring arg specify alternative built-in scoring
function: ad4_scoring default dkoes_fast
dkoes_scoring dkoes_scoring_old vina vinardo
- default/vina AutoDock Vina
- vinardo A reparameterized Vina (https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0155183)
- ad4_scoring A reimplmentation of AutoDock4 scoring (includes electrostatics and solvation)
- ignore the rest
Custom Empirical Scoring¶
Scoring functions can be defined in text files by parameterizing built-in terms
!./gnina --print_terms
electrostatic(i=2,_^=100,_c=8) ad4_solvation(d-sigma=3.6,_s/q=0.01097,_c=8) gauss(o=0,_w=0.5,_c=8) repulsion(o=0,_c=8) hydrophobic(g=0.5,_b=1.5,_c=8) non_hydrophobic(g=0.5,_b=1.5,_c=8) vdw(i=6,_j=12,_s=1,_^=100,_c=8) non_dir_h_bond_lj(o=-0.7,_^=100,_c=8) non_dir_anti_h_bond_quadratic(o=0,_c=8) non_dir_h_bond(g=-0.7,_b=0,_c=8) acceptor_acceptor_quadratic(o=0,_c=8) donor_donor_quadratic(o=0,_c=8) atom_type_gaussian(t1=,t2=,o=0,_w=0,_c=8) atom_type_linear(t1=,t2=,g=0,_b=0,_c=8) atom_type_quadratic(t1=,t2=,o=0,_c=8) atom_type_inverse_power(t1=,t2=,i=0,_^=100,_c=8) atom_type_lennard_jones(t1=,t2=,o=0,_^=100,_c=8) num_tors_add num_tors_sqr num_tors_sqrt num_tors_div num_tors_div_simple ligand_length num_ligands num_heavy_atoms_div num_heavy_atoms num_hydrophobic_atoms constant_term
Example¶
Create a file of equally-weight terms. Firt column is weight. Second the parameterized term. Remainder ignored.
open('everything.txt','wt').write('''
1.0 ad4_solvation(d-sigma=3.6,_s/q=0.01097,_c=8) desolvation, s/q is charge dependence
1.0 ad4_solvation(d-sigma=3.6,_s/q=0.0,_c=8)
1.0 electrostatic(i=1,_^=100,_c=8) i is the exponent of the distance, see everything.h for details
1.0 electrostatic(i=2,_^=100,_c=8)
1.0 gauss(o=0,_w=0.5,_c=8) o is offset, w is width of gaussian
1.0 gauss(o=3,_w=2,_c=8)
1.0 repulsion(o=0,_c=8) o is offset of squared distance repulsion
1.0 hydrophobic(g=0.5,_b=1.5,_c=8) g is a good distance, b the bad distance
1.0 non_hydrophobic(g=0.5,_b=1.5,_c=8) value is linearly interpolated between g and b
1.0 vdw(i=4,_j=8,_s=0,_^=100,_c=8) i and j are LJ exponents
1.0 vdw(i=6,_j=12,_s=1,_^=100,_c=8) s is the smoothing, ^ is the cap
1.0 non_dir_h_bond(g=-0.7,_b=0,_c=8) good and bad
1.0 non_dir_anti_h_bond_quadratic(o=0.4,_c=8) like repulsion, but for hbond, don't use
1.0 non_dir_h_bond_lj(o=-0.7,_^=100,_c=8) LJ 10-12 potential, capped at ^
1.0 acceptor_acceptor_quadratic(o=0,_c=8) quadratic potential between hydrogen bond acceptors
1.0 donor_donor_quadratic(o=0,_c=8) quadratic potential between hydroben bond donors
1.0 num_tors_div div constant terms are not linearly independent
1.0 num_heavy_atoms_div
1.0 num_heavy_atoms these terms are just added
1.0 num_tors_add
1.0 num_tors_sqr
1.0 num_tors_sqrt
1.0 num_hydrophobic_atoms
1.0 ligand_length
''');
Example¶
--custom_scoring will replace empirical scoring with function defined in provided file.
!./gnina -r rec.pdb -l lig.pdb --score_only --custom_scoring everything.txt
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec.pdb -l lig.pdb --score_only --custom_scoring everything.txt
## Name gauss(o=0,_w=0.5,_c=8) gauss(o=3,_w=2,_c=8) repulsion(o=0,_c=8) hydrophobic(g=0.5,_b=1.5,_c=8) non_hydrophobic(g=0.5,_b=1.5,_c=8) vdw(i=4,_j=8,_s=0,_^=100,_c=8) vdw(i=6,_j=12,_s=1,_^=100,_c=8) non_dir_h_bond(g=-0.7,_b=0,_c=8) non_dir_anti_h_bond_quadratic(o=0.4,_c=8) non_dir_h_bond_lj(o=-0.7,_^=100,_c=8) acceptor_acceptor_quadratic(o=0,_c=8) donor_donor_quadratic(o=0,_c=8) ad4_solvation(d-sigma=3.6,_s/q=0.01097,_c=8) ad4_solvation(d-sigma=3.6,_s/q=0,_c=8) electrostatic(i=1,_^=100,_c=8) electrostatic(i=2,_^=100,_c=8) num_tors_div num_heavy_atoms_div num_heavy_atoms num_tors_add num_tors_sqr num_tors_sqrt num_hydrophobic_atoms ligand_length
Affinity: 165.73672 (kcal/mol)
CNNscore: 0.97413
CNNaffinity: 6.98467
CNNvariance: 0.07986
Intramolecular energy: -20.40488
Term values, before weighting:
## 84.31120 1224.00134 2.82601 33.60834 56.66117 -438.07706 -498.77454 2.67216 0.24570 -18.15478 0.59442 0.00000 13.84281 -56.00148 0.18943 0.04206 0.00000 0.00000 1.25000 3.00000 0.18000 0.07746 0.45000 2.00000
Hacky Use Case¶
We wanted to soft "covalently" dock a ligand. Modified system to change atom types of bonding atoms to Chlorine and Sulfur (non-physical modification) and used this custom scoring function:
-0.035579 gauss(o=0,_w=0.5,_c=8)
-0.005156 gauss(o=3,_w=2,_c=8)
0.840245 repulsion(o=0,_c=8)
-0.035069 hydrophobic(g=0.5,_b=1.5,_c=8)
-0.587439 non_dir_h_bond(g=-0.7,_b=0,_c=8)
1.923 num_tors_div
-100.0 atom_type_gaussian(t1=Chlorine,t2=Sulfur,o=0,_w=3,_c=8)CNN Scoring¶
Convolutional neural networks learn spatially related features of an input grid to generate a prediction.
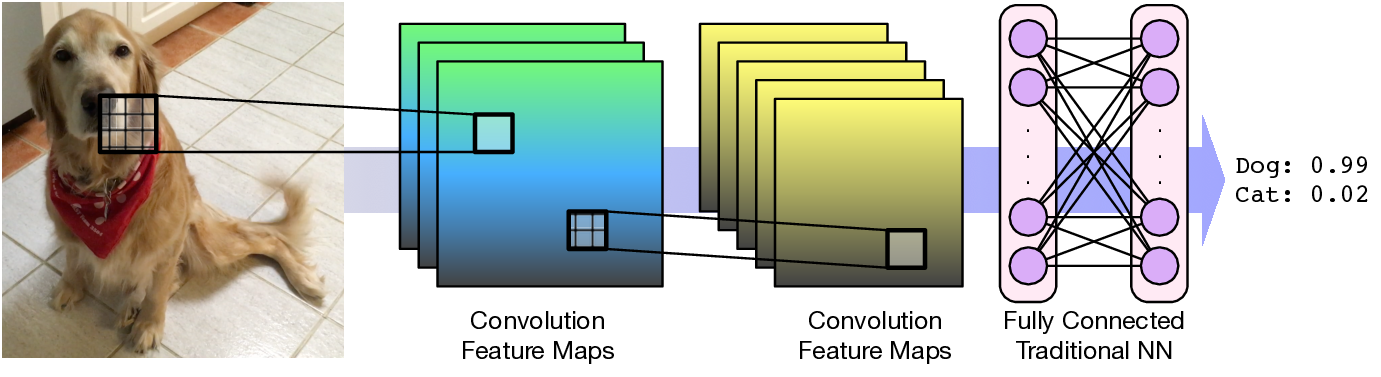
CNN Scoring¶
Atoms are represented as Gaussian densities on a 24Å grid. There is a separate channel for each atom type.
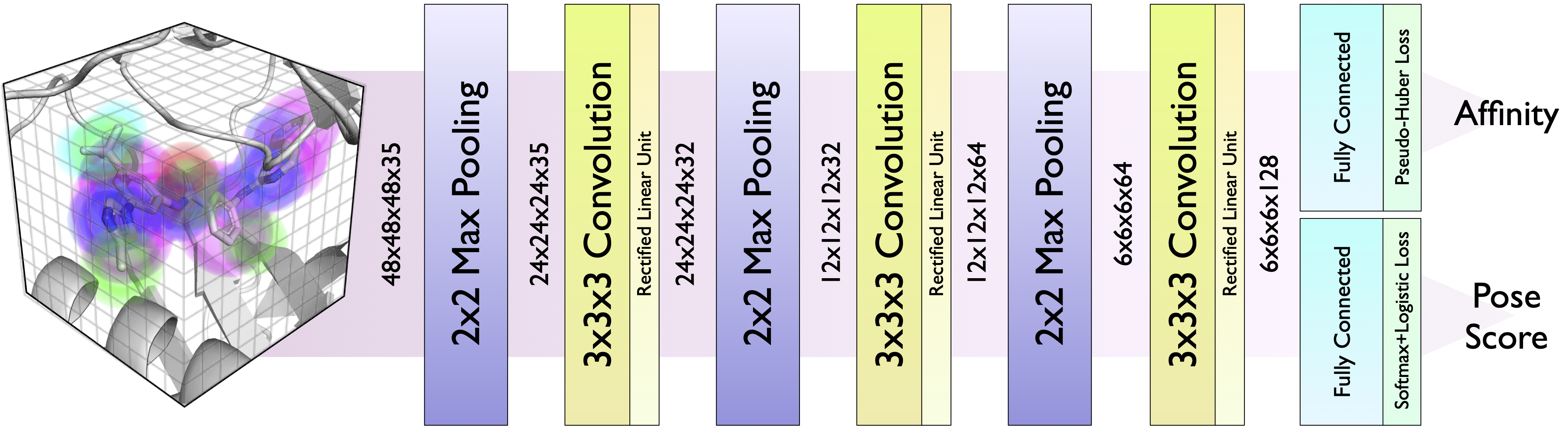
CNN Models¶
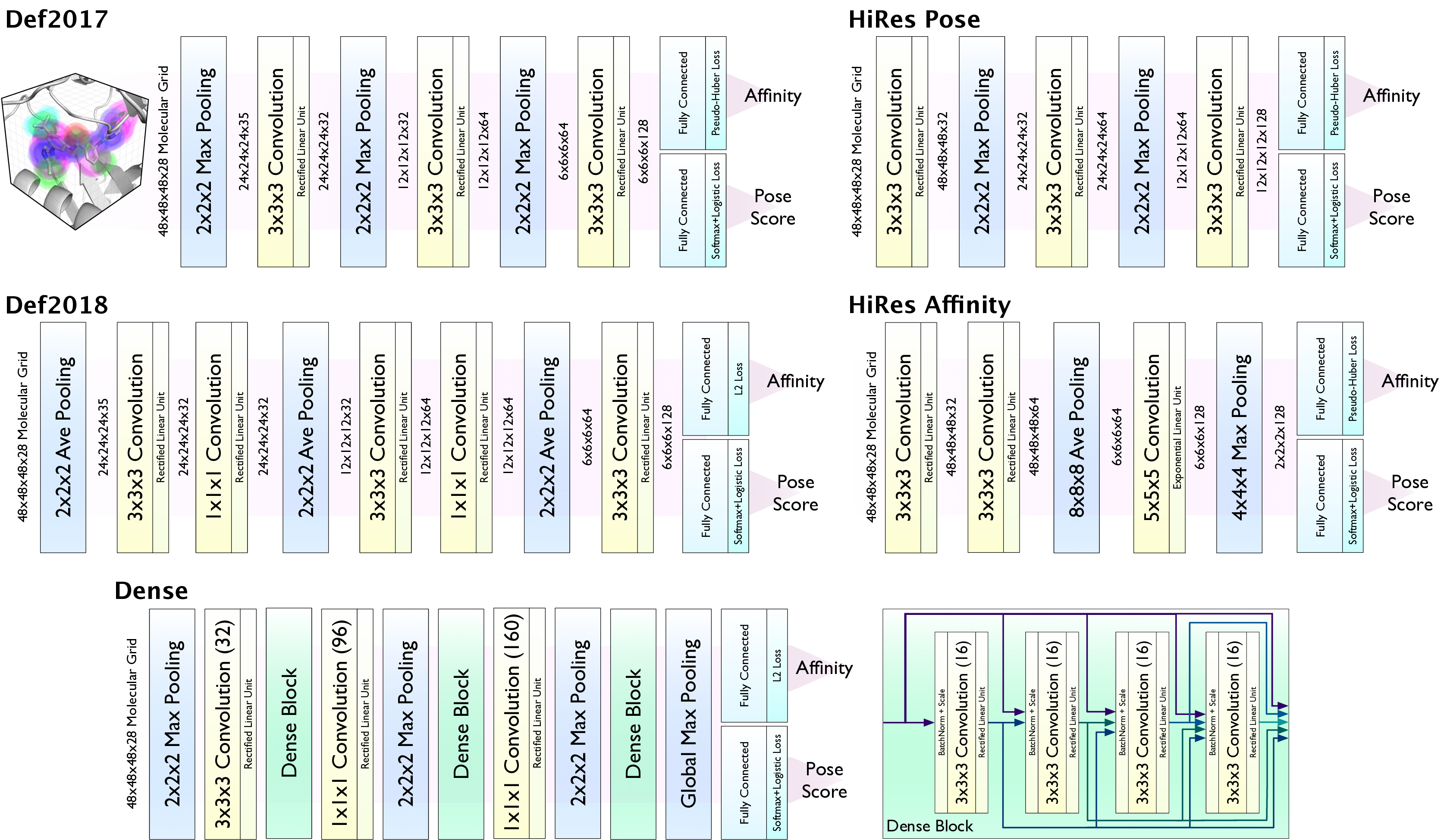
CNN Model Ensembles¶
The default is to use an ensemble of 5 models that was found to have the best performance.
!./gnina --help | grep "cnn arg" -A 12
--cnn arg built-in model to use, specify
PREFIX_ensemble to evaluate an ensemble of
models starting with PREFIX:
crossdock_default2018 crossdock_default2018_
1 crossdock_default2018_2
crossdock_default2018_3
crossdock_default2018_4 default2017 dense
dense_1 dense_2 dense_3 dense_4
general_default2018 general_default2018_1
general_default2018_2 general_default2018_3
general_default2018_4 redock_default2018
redock_default2018_1 redock_default2018_2
redock_default2018_3 redock_default2018_4
A CNN model predicts both pose quality (CNNScore) and binding affinity (CNNaffinity).
!./gnina --score_only -r rec.pdb -l lig.pdb | grep CNN
CNNscore: 0.97413 CNNaffinity: 6.98467 CNNvariance: 0.07986
CNNscore is a probability that the pose is a "good" (<2 RMSD) pose
CNNaffnity is predicted affinity in "pK" units - 1$\mu M$ is 6, 1$nM$ is 9
CNNvariance is the variance of predicted affinities across the ensemble. It is not a score, but a measure of uncertainty (lower is better).
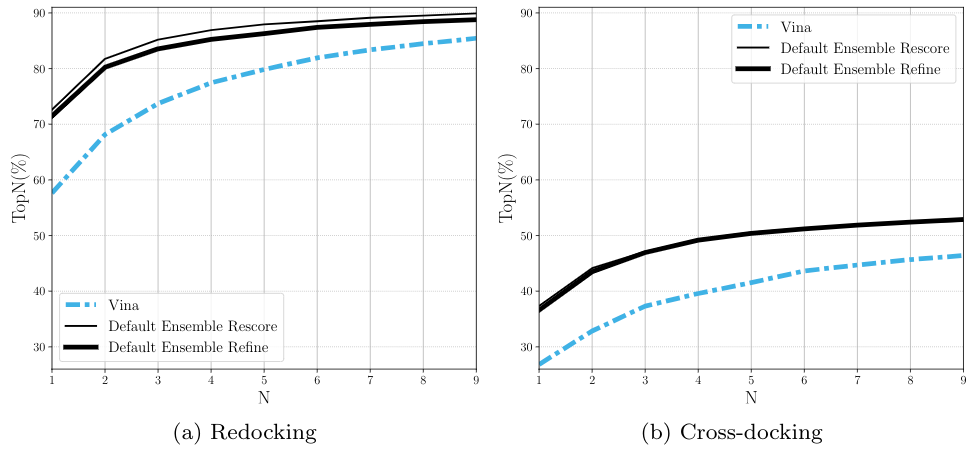
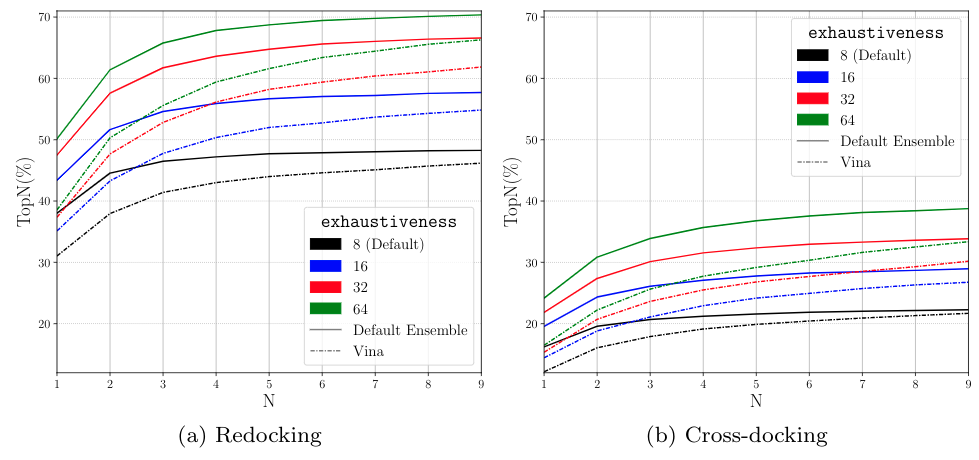
CNN Scoring Performance¶
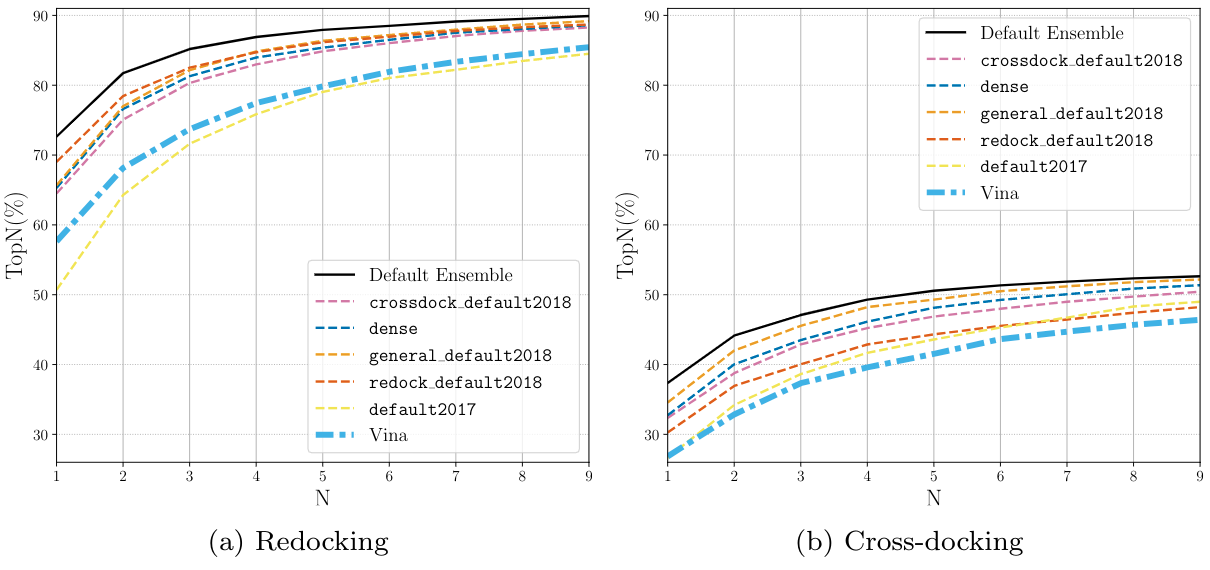
Ranking¶
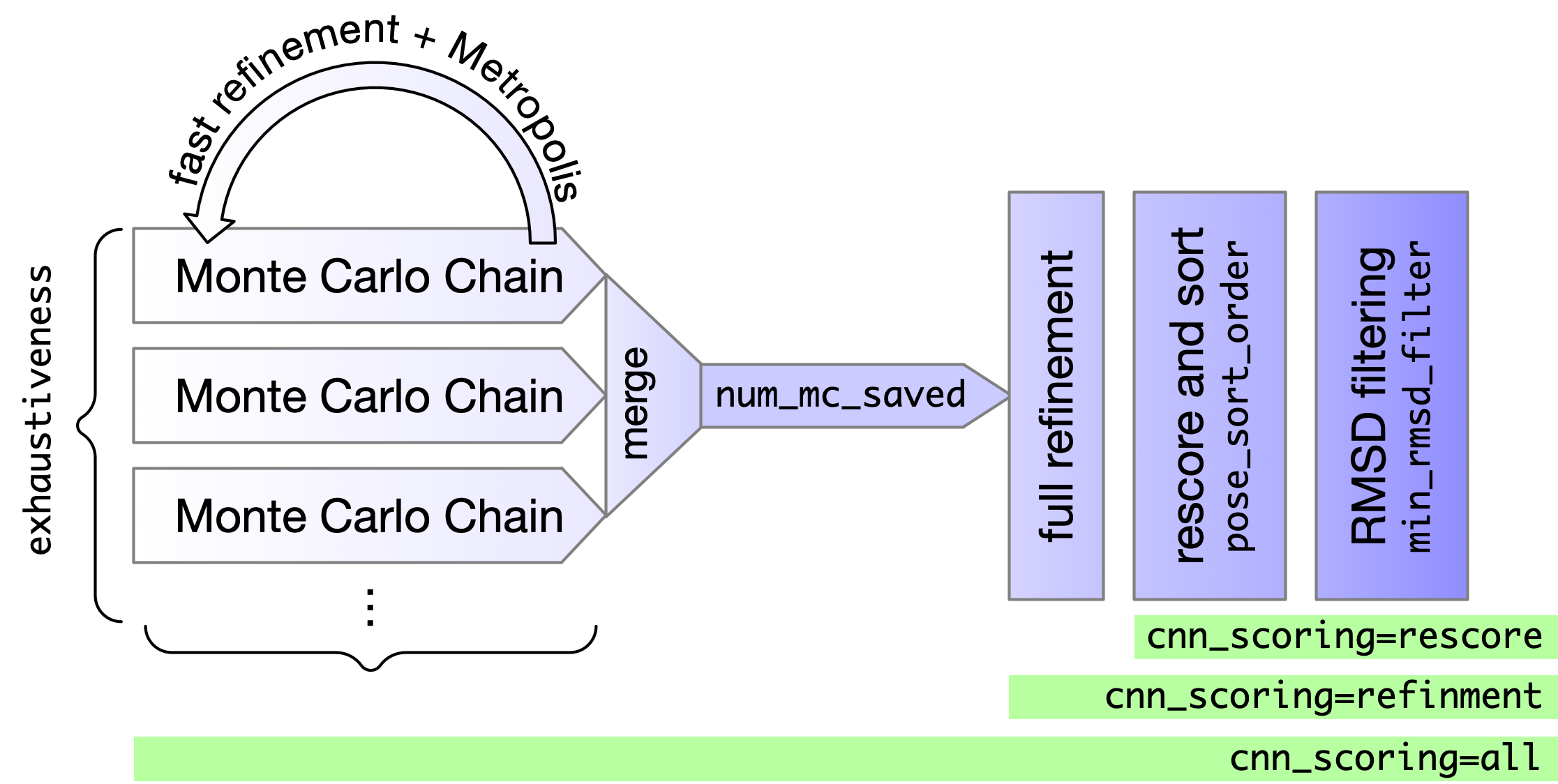
- Top
num_mc_savedposes from sampling are refined (BFGS) with full (not soft) potentials - Resulting poses are rescored and sorted according to
--pose_sort_order--pose_sort_order=CNNscore(default) Poses with highest probability of being low RMSD according to CNN are ranked highest--pose_sort_order=CNNaffinityPoses with highest CNN predicted binding affinity are ranked highest--pose_sort_order=EnergyPoses with lowest Vina predicted energy are ranked highest
- Final ranked list is filtered to remove poses within
--min_rmsd_filter(default 1Å)
Note: Changing the sort order can change what poses are returned, not just their ordering.
Using CNN for refinment (--cnn_scoring=refinement) is not helpful and is much slower.
CNN scoring is slow without a GPU. Any modern NVIDIA GPU with $\ge$4GB RAM should work.
%%time
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0 > /dev/null 2>&1
CPU times: user 107 ms, sys: 35.3 ms, total: 142 ms Wall time: 11.6 s
%%time
!CUDA_VISIBLE_DEVICES= ./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
WARNING: No GPU detected. CNN scoring will be slow.
Recommend running with single model (--cnn crossdock_default2018)
or without cnn scoring (--cnn_scoring=none).
Commandline: ./gnina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb --seed 0
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -8.52 0.9024 6.788
2 -8.09 0.6081 6.603
3 -8.31 0.4515 6.454
4 -6.62 0.3029 6.010
5 -6.24 0.2846 6.096
6 -6.83 0.2695 5.776
7 -6.86 0.1569 5.462
8 -6.76 0.1438 5.844
9 -6.16 0.1354 5.330
CPU times: user 1.67 s, sys: 299 ms, total: 1.97 s
Wall time: 2min 19s
Whole Protein Docking¶
Set the receptor to the autobox_ligand.
!./gnina -r rec.pdb -l lig.pdb --autobox_ligand rec.pdb -o wdocking.sdf.gz --seed 0
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec.pdb -l lig.pdb --autobox_ligand rec.pdb -o wdocking.sdf.gz --seed 0
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -8.49 0.8931 6.761
2 -8.08 0.6136 6.606
3 -6.52 0.4651 4.567
4 -5.40 0.3606 4.478
5 -6.02 0.3168 5.233
6 -6.88 0.2917 5.338
7 -6.44 0.2786 4.991
8 -5.24 0.2716 5.051
9 -5.61 0.2270 4.555
v = py3Dmol.view(height=400)
v.addModel(open('rec.pdb').read())
v.setStyle({'cartoon':{},'stick':{'radius':.1}})
v.addModel(open('lig.pdb').read())
v.setStyle({'model':1},{'stick':{'colorscheme':'dimgrayCarbon','radius':.125}})
v.addModelsAsFrames(gzip.open('wdocking.sdf.gz','rt').read())
v.setStyle({'model':2},{'stick':{'colorscheme':'greenCarbon'}})
v.animate({'interval':1000}); v.zoomTo(); v.rotate(90)
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944ff2df0>
We do not see diminishing returns when increasing exhaustiveness with whole protein docking.
!wget http://files.rcsb.org/download/4ERK.pdb
--2021-05-26 22:49:52-- http://files.rcsb.org/download/4ERK.pdb Resolving files.rcsb.org (files.rcsb.org)... 128.6.158.70 Connecting to files.rcsb.org (files.rcsb.org)|128.6.158.70|:80... connected. HTTP request sent, awaiting response... 200 OK Length: unspecified [application/octet-stream] Saving to: ‘4ERK.pdb’ 4ERK.pdb [ <=> ] 273.14K --.-KB/s in 0.07s 2021-05-26 22:49:52 (3.66 MB/s) - ‘4ERK.pdb’ saved [279693]
!grep ATOM 4ERK.pdb > rec2.pdb
!obabel rec2.pdb -Orec2.pdb
============================== *** Open Babel Warning in PerceiveBondOrders Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is rec2.pdb) 1 molecule converted
!grep OLO 4ERK.pdb > lig2.pdb
Let's dock the ligand from 3ERK to the 4ERK structure.
!./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o 3erk_to_4erk.sdf.gz
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o 3erk_to_4erk.sdf.gz
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -6.51 0.2157 5.896
2 -7.01 0.2154 5.674
3 -7.01 0.2018 6.019
4 -6.31 0.1858 5.833
5 -6.69 0.1834 5.585
6 -6.14 0.1785 5.550
7 -5.88 0.1611 5.485
8 -6.29 0.1555 6.021
9 -5.60 0.1465 5.313
!obrms --firstonly lig.pdb 3erk_to_4erk.sdf.gz
RMSD lig.pdb: 7.90887 RMSD lig.pdb: 5.58047 RMSD lig.pdb: 3.53731 RMSD lig.pdb: 7.67738 RMSD lig.pdb: 6.06437 RMSD lig.pdb: 7.01648 RMSD lig.pdb: 6.81812 RMSD lig.pdb: 6.42287 RMSD lig.pdb: 6.07069
v = py3Dmol.view(height=380)
v.addModel(open('3ERK.pdb').read())
v.setStyle({'model':0},{'cartoon':{'colorscheme':'greenCarbon'},'stick':{'radius':.1,'colorscheme':'greenCarbon'}})
v.addModel(open('4ERK.pdb').read())
v.setStyle({'model':1},{'cartoon':{'colorscheme':'yellowCarbon'},'stick':{'radius':.1,'colorscheme':'yellowCarbon'}})
v.addModel(gzip.open('3erk_to_4erk.sdf.gz','rt').read())
v.setStyle({'model':2},{'stick':{'colorscheme':'magentaCarbon'}})
v.zoomTo({'model':2})
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944ff5640>
Flexible Docking¶
--flexProvide flexible side-chains as PDBQT file. Rigid part of receptor should have these side-chains removed.--flexresSpecify side-chains by comma separated list of chain:resid Recommended--flexdistAll side-chains with atoms this distance fromflexdist_ligandwill be set as flexible.--flexdist_ligandLigand to use to identify side-chains by distance.--flex_limitHard limit on number of flexible residues--flex_maxSoft limit on number of flexible residues (only closest are kept)--out_flexFile to write flexible side-chain output to.makeflex.pyis provided to reassemble into full structures.
Let's try to improve our docking by making side-chains within 3Å of cognate ligand flexible.
!./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o flexdocked.sdf.gz --flexdist 4 --flexdist_ligand lig2.pdb --out_flex flexout.pdb
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o flexdocked.sdf.gz --flexdist 4 --flexdist_ligand lig2.pdb --out_flex flexout.pdb
Flexible residues: A:29 A:37 A:103 A:105 A:106 A:112 A:154
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -9.22 0.5493 6.394
2 -7.58 0.4613 6.308
3 -9.14 0.3560 6.317
4 -8.37 0.3245 6.360
5 -9.04 0.2882 6.295
6 -6.69 0.1916 5.587
7 -7.53 0.1842 5.925
8 -7.12 0.1731 5.919
9 -8.77 0.1497 6.125
!obrms --firstonly lig.pdb flexdocked.sdf.gz
RMSD lig.pdb: 2.99928 RMSD lig.pdb: 3.20943 RMSD lig.pdb: 7.221 RMSD lig.pdb: 4.97581 RMSD lig.pdb: 5.00633 RMSD lig.pdb: 3.94661 RMSD lig.pdb: 6.47123 RMSD lig.pdb: 5.59143 RMSD lig.pdb: 6.44157
v = py3Dmol.view(height=340,width=940)
v.addModel(open('3ERK.pdb').read())
v.setStyle({'model':0},{'cartoon':{'colorscheme':'greenCarbon'},'stick':{'radius':.1,'colorscheme':'greenCarbon'}})
v.addModel(open('rec2.pdb').read())
v.setStyle({'model':1},{'cartoon':{'colorscheme':'yellowCarbon'},'stick':{'radius':.1,'colorscheme':'yellowCarbon'}})
v.addModel(gzip.open('flexdocked.sdf.gz','rt').read())
v.setStyle({'model':2},{'stick':{'colorscheme':'magentaCarbon'}})
v.addModel(open('flexout.pdb').read())
v.setStyle({'model':3},{'stick':{'colorscheme':'magentaCarbon'}})
v.zoomTo({'model':2}); v.rotate(90,'x')
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944ff64f0>
Can do slightly better by being selective of what residues to make flexible and increasing exhaustiveness.
!./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o flexdocked2.sdf.gz --exhaustiveness 16 --flexres A:52,A:103 --out_flex flexout2.pdb
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r rec2.pdb -l lig.pdb --autobox_ligand lig2.pdb --seed 0 -o flexdocked2.sdf.gz --exhaustiveness 16 --flexres A:52,A:103 --out_flex flexout2.pdb
Flexible residues: A:52 A:103
Using random seed: 0
0% 10 20 30 40 50 60 70 80 90 100%
|----|----|----|----|----|----|----|----|----|----|
***************************************************
mode | affinity | CNN | CNN
| (kcal/mol) | pose score | affinity
-----+------------+------------+----------
1 -7.49 0.6000 6.474
2 -8.13 0.2974 5.994
3 -8.10 0.2610 6.179
4 -6.84 0.2601 6.143
5 -8.29 0.2056 6.233
6 -8.43 0.1907 6.018
7 -7.75 0.1887 5.962
8 -7.63 0.1756 6.288
9 -6.99 0.1740 5.980
!obrms --firstonly lig.pdb flexdocked2.sdf.gz
RMSD lig.pdb: 2.65724 RMSD lig.pdb: 4.4161 RMSD lig.pdb: 4.6292 RMSD lig.pdb: 6.99215 RMSD lig.pdb: 7.06382 RMSD lig.pdb: 7.09805 RMSD lig.pdb: 5.2716 RMSD lig.pdb: 7.20522 RMSD lig.pdb: 7.05301
v = py3Dmol.view(height=340,width=940)
v.addModel(open('3ERK.pdb').read())
v.setStyle({'model':0},{'cartoon':{'colorscheme':'greenCarbon'},'stick':{'radius':.1,'colorscheme':'greenCarbon'}})
v.addModel(open('rec2.pdb').read())
v.setStyle({'model':1},{'cartoon':{'colorscheme':'yellowCarbon'},'stick':{'radius':.1,'colorscheme':'yellowCarbon'}})
v.addModel(gzip.open('flexdocked2.sdf.gz','rt').read())
v.setStyle({'model':2},{'stick':{'colorscheme':'magentaCarbon'}})
v.addModel(open('flexout2.pdb').read())
v.setStyle({'model':3},{'stick':{'colorscheme':'magentaCarbon'}})
v.zoomTo({'model':2}); v.rotate(90,'x')
You appear to be running in JupyterLab (or JavaScript failed to load for some other reason). You need to install the 3dmol extension:
jupyter labextension install jupyterlab_3dmol
<py3Dmol.view at 0x7f3944ff5f40>
Flexible Docking Recommendations¶
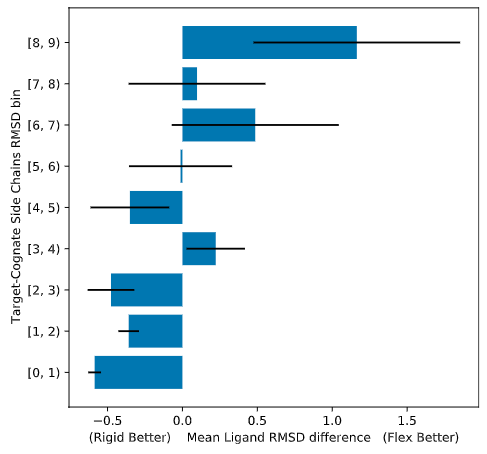
- Usually not worth it
- Increasing degrees of freedom increases false positives
- If you have an ensemble of bound protein conformations, use that
- includes backbone flexibility
- Can be useful for targetting a small number of known flexible side-chains
Virtual Screening¶
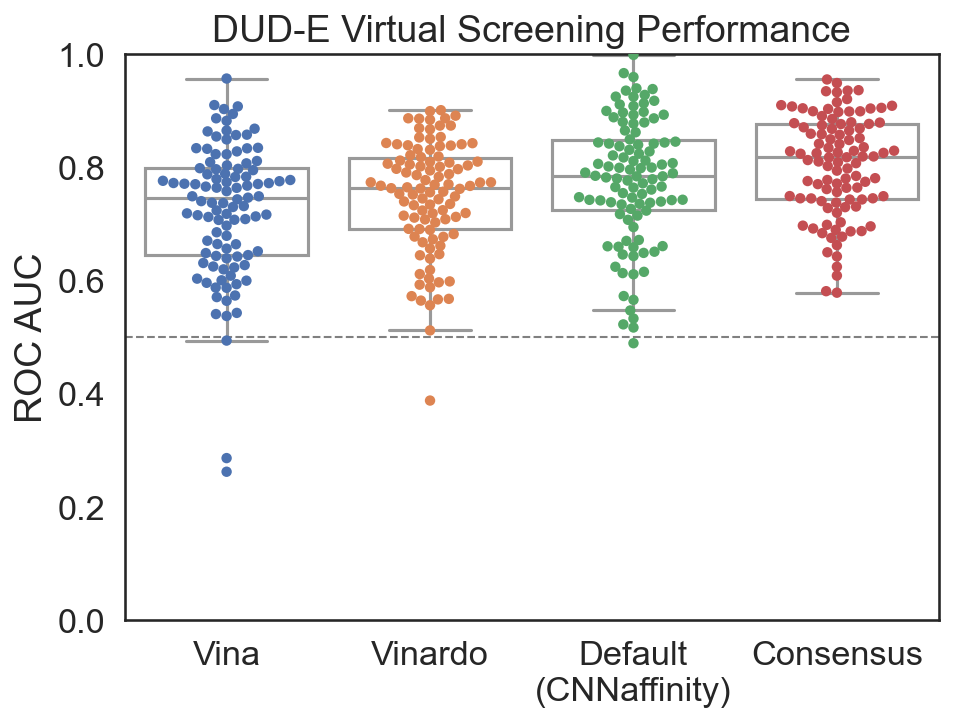
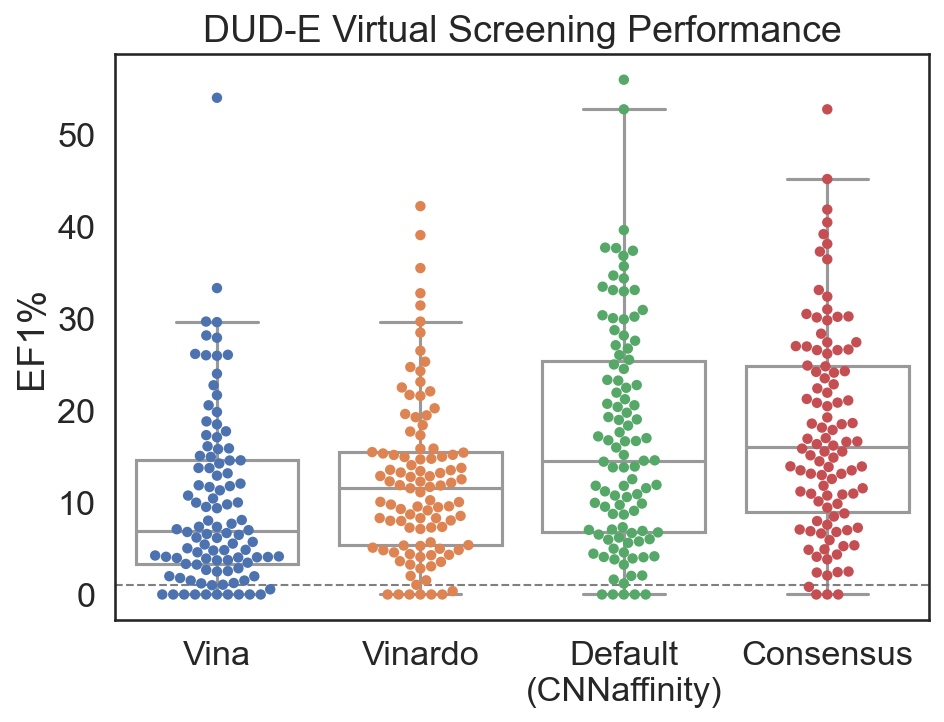
High-throughput screening recommendations¶
- Pre-filter library by molecular properties
- Remove highly flexible ligands
- Carefully manage cpu usage (
--cpu$\le$--exhaustiveness) - It is okay to share a GPU, but may be memory limited
- Avoid unnecessary receptor processing
- Provide as PDBQT
- Dock multiple ligands per a run (multi-ligand input)
- Don't do it
The Alternative: Pharmacophore Search¶
Advantages¶
- Uses expert human insight to define query
- Fast (millions of molecules in seconds)
- Results are already "docked"
- Can still using GNINA scoring to optimize/rank hits
Disadvantages¶
- Relies on expert human insight to define query
- Especially difficult when no bound ligand
- Exploration of alternative binding modes requires multiple queries
%%html
<div id="pharmexp" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#pharmexp';
jQuery(divid).asker({
id: divid,
question: "Have you done a pharmacophore search?",
answers: ['Yes w/Pharmit','Yes','No'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
Pharmit: Interactive Exploration of Chemical Space¶
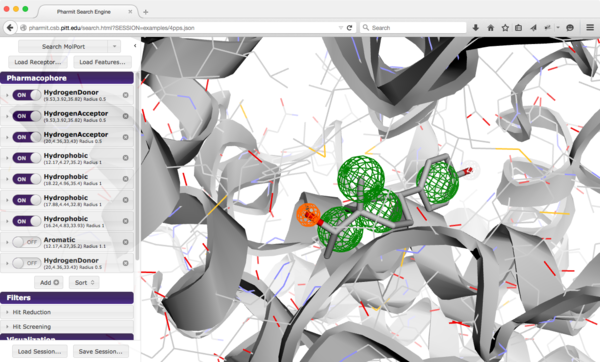
Key Points¶
- Pharmit stores rigid conformers in a special index that allows rapid pharmacophore search of millions of structures
- Several libraries with millions of commercially available compounds
- Can also create your own libraries
- Pharmacophore query specifies the 3D arrangement of essential interaction features
- Pharmacophore query specifies what must be present, not what shouldn't be
- Shape constraints can filter severe steric clashes
- Recommend minimum filtering by molecular properties
- Minimization refines ligand pose and provides Vina-based ranking
- mRMSD is how much the ligand has changed from pharmacophore-aligned pose
- large values imply no longer matches pharmacophore
- mRMSD is how much the ligand has changed from pharmacophore-aligned pose
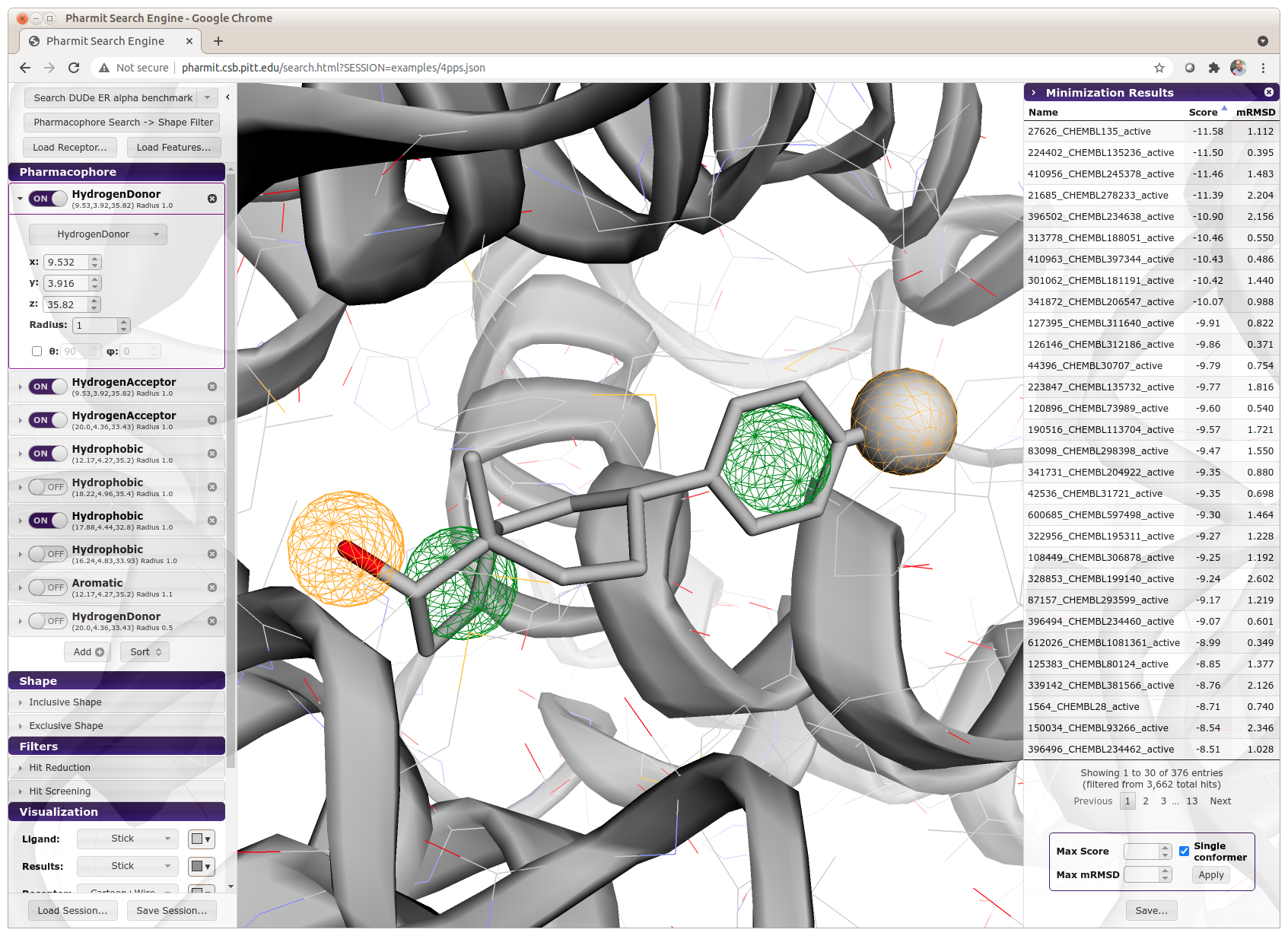
Rescoring Pharmit Results¶
Get receptor...
!wget http://files.rcsb.org/download/4PPS.pdb
--2021-05-26 22:53:41-- http://files.rcsb.org/download/4PPS.pdb Resolving files.rcsb.org (files.rcsb.org)... 128.6.158.70 Connecting to files.rcsb.org (files.rcsb.org)|128.6.158.70|:80... connected. HTTP request sent, awaiting response... 200 OK Length: unspecified [application/octet-stream] Saving to: ‘4PPS.pdb’ 4PPS.pdb [ <=> ] 721.01K --.-KB/s in 0.1s 2021-05-26 22:53:41 (6.47 MB/s) - ‘4PPS.pdb’ saved [738315]
!grep ^ATOM 4PPS.pdb > errec.pdb
Rescoring Pharmit Results¶
!./gnina -r errec.pdb -l minimized_results.sdf.gz --minimize -o gnina_scored.sdf.gz --scoring vinardo
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina v1.0.1 HEAD:aa41230 Built Mar 23 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: ./gnina -r errec.pdb -l minimized_results.sdf.gz --minimize -o gnina_scored.sdf.gz --scoring vinardo
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders
Affinity: -9.72913 0.00000 (kcal/mol)
RMSD: 0.26195
CNNscore: 0.89913
CNNaffinity: 7.91113
CNNvariance: 0.65732
Affinity: -9.20967 -0.70125 (kcal/mol)
RMSD: 0.31283
CNNscore: 0.85194
CNNaffinity: 7.48067
CNNvariance: 0.45972
Affinity: -8.73188 0.00000 (kcal/mol)
RMSD: 0.14765
CNNscore: 0.61851
CNNaffinity: 7.48549
CNNvariance: 0.08808
Affinity: -9.65893 0.24209 (kcal/mol)
RMSD: 0.33742
CNNscore: 0.94762
CNNaffinity: 7.80676
CNNvariance: 0.81808
Affinity: -9.80264 0.00000 (kcal/mol)
RMSD: 0.64717
CNNscore: 0.93777
CNNaffinity: 7.64913
CNNvariance: 0.31171
Affinity: -8.81335 0.00000 (kcal/mol)
RMSD: 0.20502
CNNscore: 0.86194
CNNaffinity: 7.89758
CNNvariance: 0.74401
Affinity: -7.51991 0.00000 (kcal/mol)
RMSD: 0.17124
CNNscore: 0.81780
CNNaffinity: 7.94833
CNNvariance: 0.38997
Affinity: -8.25420 -1.02751 (kcal/mol)
RMSD: 0.26137
CNNscore: 0.95250
CNNaffinity: 7.65842
CNNvariance: 0.06438
Affinity: -8.40613 0.07375 (kcal/mol)
RMSD: 0.21762
CNNscore: 0.89652
CNNaffinity: 7.61298
CNNvariance: 0.70283
Affinity: -8.89704 0.00000 (kcal/mol)
RMSD: 0.40716
CNNscore: 0.49027
CNNaffinity: 6.90353
CNNvariance: 0.29763
Affinity: -9.02506 0.00000 (kcal/mol)
RMSD: 0.53020
CNNscore: 0.63625
CNNaffinity: 6.89807
CNNvariance: 0.16616
Affinity: -8.08758 0.00000 (kcal/mol)
RMSD: 0.13200
CNNscore: 0.84075
CNNaffinity: 7.19361
CNNvariance: 0.45240
Affinity: -8.19398 0.00000 (kcal/mol)
RMSD: 0.27467
CNNscore: 0.92643
CNNaffinity: 7.76735
CNNvariance: 0.68770
Affinity: -8.09598 0.00000 (kcal/mol)
RMSD: 0.19189
CNNscore: 0.72409
CNNaffinity: 7.18069
CNNvariance: 1.22493
Affinity: -8.24613 -0.57158 (kcal/mol)
RMSD: 0.13589
CNNscore: 0.83741
CNNaffinity: 7.18732
CNNvariance: 0.67518
Affinity: -6.91419 0.00000 (kcal/mol)
RMSD: 0.48355
CNNscore: 0.62871
CNNaffinity: 6.34572
CNNvariance: 0.44166
Affinity: -7.53146 0.00000 (kcal/mol)
RMSD: 0.19259
CNNscore: 0.67640
CNNaffinity: 6.85396
CNNvariance: 0.20357
Affinity: -8.40987 -0.06287 (kcal/mol)
RMSD: 0.13818
CNNscore: 0.76788
CNNaffinity: 7.17220
CNNvariance: 0.41133
Affinity: -4.73829 0.06123 (kcal/mol)
RMSD: 0.21484
CNNscore: 0.37701
CNNaffinity: 7.64041
CNNvariance: 0.34767
Affinity: -9.01593 -0.63920 (kcal/mol)
RMSD: 0.13379
CNNscore: 0.84997
CNNaffinity: 7.37605
CNNvariance: 0.56718
Affinity: -9.54805 -0.21692 (kcal/mol)
RMSD: 0.58113
CNNscore: 0.83030
CNNaffinity: 7.75141
CNNvariance: 0.54246
Affinity: -8.57497 -1.82244 (kcal/mol)
RMSD: 0.59535
CNNscore: 0.59916
CNNaffinity: 7.77063
CNNvariance: 0.57655
Affinity: -8.50449 -0.41720 (kcal/mol)
RMSD: 0.16117
CNNscore: 0.86746
CNNaffinity: 7.35945
CNNvariance: 1.30343
Affinity: -8.38439 -0.43862 (kcal/mol)
RMSD: 0.08202
CNNscore: 0.80857
CNNaffinity: 7.00821
CNNvariance: 0.59343
Affinity: -7.35425 0.00000 (kcal/mol)
RMSD: 0.29391
CNNscore: 0.27216
CNNaffinity: 7.10966
CNNvariance: 0.48634
Affinity: -7.89201 -0.69650 (kcal/mol)
RMSD: 0.26468
CNNscore: 0.76073
CNNaffinity: 7.67225
CNNvariance: 0.76690
Affinity: -6.53450 0.13349 (kcal/mol)
RMSD: 0.18472
CNNscore: 0.60232
CNNaffinity: 7.82335
CNNvariance: 0.49484
Affinity: -8.07613 -0.39550 (kcal/mol)
RMSD: 0.34820
CNNscore: 0.66062
CNNaffinity: 7.05296
CNNvariance: 0.82901
Affinity: -7.09084 -0.40356 (kcal/mol)
RMSD: 0.64751
CNNscore: 0.62292
CNNaffinity: 7.49378
CNNvariance: 0.87468
Affinity: -7.31595 -0.66102 (kcal/mol)
RMSD: 0.89548
CNNscore: 0.57088
CNNaffinity: 7.47125
CNNvariance: 1.14700
Affinity: -6.00163 -0.21792 (kcal/mol)
RMSD: 0.18096
CNNscore: 0.54068
CNNaffinity: 7.95480
CNNvariance: 0.70406
Affinity: -7.93454 -0.12205 (kcal/mol)
RMSD: 0.60941
CNNscore: 0.74976
CNNaffinity: 7.19810
CNNvariance: 0.86179
Affinity: -8.36049 -0.19293 (kcal/mol)
RMSD: 0.45915
CNNscore: 0.65698
CNNaffinity: 6.90297
CNNvariance: 0.54429
Affinity: -7.31305 0.11529 (kcal/mol)
RMSD: 0.30822
CNNscore: 0.77150
CNNaffinity: 7.26929
CNNvariance: 0.47337
Affinity: -6.72021 -0.59378 (kcal/mol)
RMSD: 0.19239
CNNscore: 0.55537
CNNaffinity: 7.36714
CNNvariance: 0.43517
Affinity: -8.55884 -0.38827 (kcal/mol)
RMSD: 0.60655
CNNscore: 0.84384
CNNaffinity: 7.67851
CNNvariance: 0.81399
Affinity: -6.99289 -0.04329 (kcal/mol)
RMSD: 0.82451
CNNscore: 0.72689
CNNaffinity: 7.38528
CNNvariance: 0.65457
Affinity: -5.97045 -0.62302 (kcal/mol)
RMSD: 0.31094
CNNscore: 0.51557
CNNaffinity: 7.21088
CNNvariance: 0.82040
Affinity: -4.89224 -0.63006 (kcal/mol)
RMSD: 0.15241
CNNscore: 0.15759
CNNaffinity: 7.72342
CNNvariance: 0.19575
Affinity: -6.85474 -0.63239 (kcal/mol)
RMSD: 0.18048
CNNscore: 0.58627
CNNaffinity: 6.97516
CNNvariance: 0.78292
Affinity: -6.53305 -0.29451 (kcal/mol)
RMSD: 0.51028
CNNscore: 0.56192
CNNaffinity: 6.99936
CNNvariance: 0.85128
Affinity: -7.23467 -0.76411 (kcal/mol)
RMSD: 1.11674
CNNscore: 0.46142
CNNaffinity: 6.79067
CNNvariance: 0.34035
Affinity: -7.04893 -0.39319 (kcal/mol)
RMSD: 0.62040
CNNscore: 0.76078
CNNaffinity: 7.16045
CNNvariance: 0.58269
Affinity: -6.83654 0.37302 (kcal/mol)
RMSD: 0.35913
CNNscore: 0.50458
CNNaffinity: 7.33000
CNNvariance: 0.03670
Affinity: -6.56503 -0.38408 (kcal/mol)
RMSD: 0.20366
CNNscore: 0.72248
CNNaffinity: 7.39523
CNNvariance: 1.02604
Affinity: -6.55665 -1.13885 (kcal/mol)
RMSD: 0.25327
CNNscore: 0.57688
CNNaffinity: 7.36374
CNNvariance: 0.86582
Affinity: -5.74291 -0.61074 (kcal/mol)
RMSD: 0.23124
CNNscore: 0.28129
CNNaffinity: 7.24846
CNNvariance: 0.40314
Affinity: -4.94074 -0.78787 (kcal/mol)
RMSD: 0.74550
CNNscore: 0.51484
CNNaffinity: 7.51896
CNNvariance: 1.24439
Affinity: -6.02883 -1.35000 (kcal/mol)
RMSD: 0.09003
CNNscore: 0.19070
CNNaffinity: 6.98689
CNNvariance: 0.63347
Affinity: -4.12884 -0.11860 (kcal/mol)
RMSD: 0.30044
CNNscore: 0.45804
CNNaffinity: 7.23205
CNNvariance: 0.17427
Affinity: -3.50053 1.68315 (kcal/mol)
RMSD: 0.14996
CNNscore: 0.50196
CNNaffinity: 7.83518
CNNvariance: 0.37307
Affinity: -5.06636 -0.33127 (kcal/mol)
RMSD: 0.22011
CNNscore: 0.15402
CNNaffinity: 7.44639
CNNvariance: 0.08817
Affinity: -8.01032 -0.23865 (kcal/mol)
RMSD: 0.70319
CNNscore: 0.58079
CNNaffinity: 7.11340
CNNvariance: 0.78848
Affinity: -4.79429 -2.01972 (kcal/mol)
RMSD: 0.26305
CNNscore: 0.50249
CNNaffinity: 7.74883
CNNvariance: 0.58747
Affinity: -6.67288 -1.61712 (kcal/mol)
RMSD: 0.40466
CNNscore: 0.32523
CNNaffinity: 7.23326
CNNvariance: 0.60915
Affinity: -5.57535 0.00292 (kcal/mol)
RMSD: 0.41003
CNNscore: 0.43508
CNNaffinity: 6.55695
CNNvariance: 0.26199
Affinity: -5.95157 -0.85276 (kcal/mol)
RMSD: 0.24428
CNNscore: 0.35815
CNNaffinity: 6.41227
CNNvariance: 0.12377
Affinity: -4.66424 -0.79291 (kcal/mol)
RMSD: 0.65341
CNNscore: 0.36316
CNNaffinity: 7.18383
CNNvariance: 0.27606
Affinity: -4.44709 -0.41284 (kcal/mol)
RMSD: 0.11685
CNNscore: 0.23159
CNNaffinity: 7.22095
CNNvariance: 0.17717
Affinity: -4.70860 -0.30222 (kcal/mol)
RMSD: 0.13248
CNNscore: 0.51042
CNNaffinity: 7.14567
CNNvariance: 0.84602
Affinity: -4.84197 -0.07825 (kcal/mol)
RMSD: 0.77146
CNNscore: 0.40603
CNNaffinity: 7.05291
CNNvariance: 0.65008
Affinity: -4.15430 -0.64001 (kcal/mol)
RMSD: 0.06970
CNNscore: 0.51594
CNNaffinity: 7.31764
CNNvariance: 1.47518
Affinity: -4.55380 0.89607 (kcal/mol)
RMSD: 0.24656
CNNscore: 0.15415
CNNaffinity: 7.24488
CNNvariance: 0.19297
Affinity: -6.51563 -1.17947 (kcal/mol)
RMSD: 0.69327
CNNscore: 0.22701
CNNaffinity: 6.92217
CNNvariance: 0.16619
Affinity: -0.57789 1.66126 (kcal/mol)
RMSD: 0.11632
CNNscore: 0.08574
CNNaffinity: 6.81906
CNNvariance: 0.26483
Affinity: -2.34578 -2.26052 (kcal/mol)
RMSD: 0.10379
CNNscore: 0.24558
CNNaffinity: 7.53951
CNNvariance: 0.15278
Affinity: -3.05835 -0.21120 (kcal/mol)
RMSD: 0.17273
CNNscore: 0.09575
CNNaffinity: 7.09038
CNNvariance: 0.22568
Affinity: -3.04384 -0.86395 (kcal/mol)
RMSD: 0.15464
CNNscore: 0.11432
CNNaffinity: 7.10941
CNNvariance: 0.26608
Affinity: -2.00272 -0.28177 (kcal/mol)
RMSD: 0.22866
CNNscore: 0.08859
CNNaffinity: 6.91712
CNNvariance: 0.39314
Affinity: -0.88033 2.11105 (kcal/mol)
RMSD: 0.18261
CNNscore: 0.13971
CNNaffinity: 7.15275
CNNvariance: 0.66823
Affinity: -5.31016 -1.05632 (kcal/mol)
RMSD: 1.45562
CNNscore: 0.11628
CNNaffinity: 6.97752
CNNvariance: 0.28533
Affinity: -1.41147 -0.91734 (kcal/mol)
RMSD: 0.22343
CNNscore: 0.12788
CNNaffinity: 7.30785
CNNvariance: 0.10973
Affinity: -2.16153 -0.82381 (kcal/mol)
RMSD: 1.58751
CNNscore: 0.52175
CNNaffinity: 7.34284
CNNvariance: 0.42344
Affinity: -3.01033 -0.39341 (kcal/mol)
RMSD: 0.33445
CNNscore: 0.48155
CNNaffinity: 7.12270
CNNvariance: 0.78472
Affinity: -5.01017 -0.95675 (kcal/mol)
RMSD: 0.81920
CNNscore: 0.36479
CNNaffinity: 6.91952
CNNvariance: 0.25837
Affinity: -1.14432 -1.10631 (kcal/mol)
RMSD: 0.17286
CNNscore: 0.13926
CNNaffinity: 7.44877
CNNvariance: 0.19926
Affinity: -2.01584 -1.53158 (kcal/mol)
RMSD: 0.12354
CNNscore: 0.31457
CNNaffinity: 7.12730
CNNvariance: 1.00547
Affinity: 0.33789 -2.99404 (kcal/mol)
RMSD: 0.18828
CNNscore: 0.22065
CNNaffinity: 7.57260
CNNvariance: 0.66164
Affinity: -0.72968 -0.08745 (kcal/mol)
RMSD: 0.43902
CNNscore: 0.25283
CNNaffinity: 7.63603
CNNvariance: 0.10144
Affinity: -1.23333 -1.84437 (kcal/mol)
RMSD: 0.12354
CNNscore: 0.12490
CNNaffinity: 7.55654
CNNvariance: 0.15117
Affinity: 0.27515 -2.14841 (kcal/mol)
RMSD: 0.08431
CNNscore: 0.07607
CNNaffinity: 6.49941
CNNvariance: 0.49137
Affinity: -0.12303 -0.06943 (kcal/mol)
RMSD: 0.12695
CNNscore: 0.61448
CNNaffinity: 7.92108
CNNvariance: 0.48226
Affinity: -0.38321 -1.43049 (kcal/mol)
RMSD: 1.12721
CNNscore: 0.28607
CNNaffinity: 7.70992
CNNvariance: 0.59401
Affinity: -0.76925 -1.40123 (kcal/mol)
RMSD: 0.25604
CNNscore: 0.32888
CNNaffinity: 7.09082
CNNvariance: 0.35006
Affinity: 0.72094 0.51718 (kcal/mol)
RMSD: 0.37653
CNNscore: 0.21924
CNNaffinity: 7.51318
CNNvariance: 0.76549
Affinity: -1.30457 -3.20178 (kcal/mol)
RMSD: 0.31826
CNNscore: 0.53165
CNNaffinity: 7.78917
CNNvariance: 0.48310
Affinity: -1.99333 -0.48469 (kcal/mol)
RMSD: 0.52436
CNNscore: 0.52990
CNNaffinity: 7.44349
CNNvariance: 0.36006
Affinity: -3.07390 -0.69669 (kcal/mol)
RMSD: 1.28659
CNNscore: 0.13340
CNNaffinity: 7.18738
CNNvariance: 0.93055
Affinity: 1.24017 -1.34054 (kcal/mol)
RMSD: 0.14647
CNNscore: 0.06432
CNNaffinity: 6.72039
CNNvariance: 0.10788
Affinity: -1.78209 -1.17221 (kcal/mol)
RMSD: 0.14907
CNNscore: 0.37828
CNNaffinity: 7.71119
CNNvariance: 1.92875
Affinity: -2.92270 -0.18884 (kcal/mol)
RMSD: 0.21510
CNNscore: 0.40707
CNNaffinity: 7.46790
CNNvariance: 0.36607
Affinity: -1.89068 -1.64813 (kcal/mol)
RMSD: 0.48571
CNNscore: 0.34864
CNNaffinity: 7.27456
CNNvariance: 0.81913
Affinity: 0.41155 -2.07781 (kcal/mol)
RMSD: 0.15981
CNNscore: 0.12504
CNNaffinity: 7.09032
CNNvariance: 0.24951
Affinity: -0.54734 -1.33615 (kcal/mol)
RMSD: 0.11486
CNNscore: 0.15381
CNNaffinity: 7.21677
CNNvariance: 0.14212
Affinity: 0.65948 -4.07207 (kcal/mol)
RMSD: 0.16546
CNNscore: 0.06565
CNNaffinity: 7.18832
CNNvariance: 0.45164
Affinity: 0.48295 -0.67816 (kcal/mol)
RMSD: 0.15494
CNNscore: 0.14528
CNNaffinity: 7.51533
CNNvariance: 0.25769
Affinity: 0.57203 -0.77223 (kcal/mol)
RMSD: 0.10361
CNNscore: 0.15437
CNNaffinity: 7.47899
CNNvariance: 0.27103
Affinity: 0.71760 -0.55153 (kcal/mol)
RMSD: 0.44994
CNNscore: 0.24544
CNNaffinity: 6.74337
CNNvariance: 0.38176
Affinity: 0.41284 -0.47117 (kcal/mol)
RMSD: 0.27232
CNNscore: 0.45073
CNNaffinity: 6.95680
CNNvariance: 1.14258
Affinity: 1.21045 -3.78420 (kcal/mol)
RMSD: 0.15185
CNNscore: 0.44033
CNNaffinity: 7.68483
CNNvariance: 0.67297
Affinity: 3.20431 -0.67891 (kcal/mol)
RMSD: 0.55638
CNNscore: 0.19058
CNNaffinity: 7.31367
CNNvariance: 0.39536
Affinity: 2.02540 -0.84809 (kcal/mol)
RMSD: 0.39304
CNNscore: 0.38106
CNNaffinity: 7.44568
CNNvariance: 0.39586
Affinity: 3.29499 0.28763 (kcal/mol)
RMSD: 0.40616
CNNscore: 0.30761
CNNaffinity: 7.85613
CNNvariance: 0.13245
Affinity: 1.32021 -1.19424 (kcal/mol)
RMSD: 1.04855
CNNscore: 0.11796
CNNaffinity: 7.13823
CNNvariance: 0.31093
Affinity: -0.05997 -0.27305 (kcal/mol)
RMSD: 0.99394
CNNscore: 0.39303
CNNaffinity: 7.19539
CNNvariance: 0.49764
Affinity: 0.96975 0.01511 (kcal/mol)
RMSD: 1.14594
CNNscore: 0.07669
CNNaffinity: 6.96628
CNNvariance: 0.16378
Affinity: 3.36695 -1.28348 (kcal/mol)
RMSD: 0.42188
CNNscore: 0.43187
CNNaffinity: 7.81816
CNNvariance: 0.68962
Affinity: 2.76042 -0.26968 (kcal/mol)
RMSD: 0.23257
CNNscore: 0.25707
CNNaffinity: 7.78630
CNNvariance: 0.29916
Affinity: 6.44258 -0.79150 (kcal/mol)
RMSD: 0.13227
CNNscore: 0.25127
CNNaffinity: 7.07448
CNNvariance: 0.29959
Affinity: 0.45507 -0.21126 (kcal/mol)
RMSD: 0.11427
CNNscore: 0.41971
CNNaffinity: 8.01693
CNNvariance: 0.27126
Affinity: 2.14339 -1.36472 (kcal/mol)
RMSD: 1.20782
CNNscore: 0.07320
CNNaffinity: 7.11523
CNNvariance: 0.63003
Affinity: 1.75388 -1.08391 (kcal/mol)
RMSD: 0.46042
CNNscore: 0.06499
CNNaffinity: 7.33678
CNNvariance: 0.24067
Affinity: 1.53301 -1.74144 (kcal/mol)
RMSD: 0.19163
CNNscore: 0.27878
CNNaffinity: 7.81152
CNNvariance: 0.30706
Affinity: 0.59089 -0.40748 (kcal/mol)
RMSD: 0.17161
CNNscore: 0.20872
CNNaffinity: 7.79298
CNNvariance: 0.20135
Affinity: 3.06404 -0.71275 (kcal/mol)
RMSD: 0.17986
CNNscore: 0.49220
CNNaffinity: 7.89068
CNNvariance: 0.69491
Affinity: 1.06985 -0.44324 (kcal/mol)
RMSD: 0.23674
CNNscore: 0.41894
CNNaffinity: 7.25090
CNNvariance: 0.35501
Affinity: 2.98184 -1.50641 (kcal/mol)
RMSD: 0.25230
CNNscore: 0.53258
CNNaffinity: 7.67904
CNNvariance: 0.68862
Affinity: 2.00456 -1.02162 (kcal/mol)
RMSD: 0.20821
CNNscore: 0.43185
CNNaffinity: 7.88471
CNNvariance: 1.59273
Affinity: 2.62157 -0.70057 (kcal/mol)
RMSD: 0.53156
CNNscore: 0.20734
CNNaffinity: 7.08090
CNNvariance: 0.17169
Affinity: 3.24824 -0.03359 (kcal/mol)
RMSD: 0.09693
CNNscore: 0.03747
CNNaffinity: 7.51142
CNNvariance: 0.03042
Affinity: 2.55078 1.34559 (kcal/mol)
RMSD: 1.64608
CNNscore: 0.36757
CNNaffinity: 7.63379
CNNvariance: 0.17979
Affinity: 2.99709 -2.05661 (kcal/mol)
RMSD: 0.78159
CNNscore: 0.33823
CNNaffinity: 7.94940
CNNvariance: 0.48080
Affinity: 2.30901 -1.01653 (kcal/mol)
RMSD: 0.23543
CNNscore: 0.24447
CNNaffinity: 7.67116
CNNvariance: 0.32847
Affinity: 3.58967 -1.66613 (kcal/mol)
RMSD: 0.11579
CNNscore: 0.09078
CNNaffinity: 7.45779
CNNvariance: 0.56416
Affinity: 5.50731 0.61115 (kcal/mol)
RMSD: 0.21714
CNNscore: 0.14898
CNNaffinity: 7.41831
CNNvariance: 0.49967
Affinity: 2.64835 -1.90108 (kcal/mol)
RMSD: 0.15390
CNNscore: 0.15728
CNNaffinity: 7.40782
CNNvariance: 0.05617
Affinity: 2.73176 -0.22138 (kcal/mol)
RMSD: 0.12009
CNNscore: 0.51223
CNNaffinity: 6.94588
CNNvariance: 1.08259
Affinity: 4.22078 -0.87711 (kcal/mol)
RMSD: 0.12988
CNNscore: 0.15257
CNNaffinity: 7.31820
CNNvariance: 0.87672
Affinity: 3.45628 -0.48729 (kcal/mol)
RMSD: 0.21663
CNNscore: 0.34749
CNNaffinity: 7.36222
CNNvariance: 0.42377
Affinity: 2.40440 -1.34474 (kcal/mol)
RMSD: 0.32391
CNNscore: 0.22717
CNNaffinity: 7.01356
CNNvariance: 0.29402
Affinity: 3.83889 -3.13488 (kcal/mol)
RMSD: 0.16967
CNNscore: 0.22396
CNNaffinity: 7.29419
CNNvariance: 0.21676
Affinity: 3.18921 -3.00389 (kcal/mol)
RMSD: 0.31746
CNNscore: 0.09140
CNNaffinity: 7.09559
CNNvariance: 0.06709
Affinity: 3.24217 -1.47823 (kcal/mol)
RMSD: 0.21903
CNNscore: 0.55389
CNNaffinity: 7.67133
CNNvariance: 0.25801
Affinity: 3.61688 -1.08076 (kcal/mol)
RMSD: 0.09962
CNNscore: 0.04354
CNNaffinity: 7.13083
CNNvariance: 0.08458
Affinity: 5.01546 0.91840 (kcal/mol)
RMSD: 0.10509
CNNscore: 0.21124
CNNaffinity: 7.47121
CNNvariance: 0.31809
Affinity: 1.02278 -0.16015 (kcal/mol)
RMSD: 1.80571
CNNscore: 0.14962
CNNaffinity: 7.33922
CNNvariance: 0.18787
Affinity: 3.86801 0.86974 (kcal/mol)
RMSD: 0.17703
CNNscore: 0.10519
CNNaffinity: 7.22803
CNNvariance: 0.06891
Affinity: 4.84045 0.22221 (kcal/mol)
RMSD: 0.09558
CNNscore: 0.19994
CNNaffinity: 7.46969
CNNvariance: 0.12534
Affinity: 3.33705 -1.14187 (kcal/mol)
RMSD: 0.12437
CNNscore: 0.20141
CNNaffinity: 7.52294
CNNvariance: 0.20242
Affinity: 4.27317 -1.64062 (kcal/mol)
RMSD: 0.19943
CNNscore: 0.50813
CNNaffinity: 7.74929
CNNvariance: 0.44028
Affinity: 3.79317 1.73598 (kcal/mol)
RMSD: 0.17359
CNNscore: 0.28110
CNNaffinity: 7.66049
CNNvariance: 0.22407
Affinity: 2.02095 -1.15973 (kcal/mol)
RMSD: 0.89729
CNNscore: 0.56386
CNNaffinity: 7.64444
CNNvariance: 0.78097
Affinity: 3.82778 0.12033 (kcal/mol)
RMSD: 0.11531
CNNscore: 0.24635
CNNaffinity: 7.18424
CNNvariance: 0.35501
Affinity: 4.80275 -1.35223 (kcal/mol)
RMSD: 0.41837
CNNscore: 0.10761
CNNaffinity: 7.24550
CNNvariance: 0.44100
Affinity: -2.23867 -0.90913 (kcal/mol)
RMSD: 1.23259
CNNscore: 0.12923
CNNaffinity: 7.08193
CNNvariance: 0.14523
Affinity: 4.61102 -0.67283 (kcal/mol)
RMSD: 0.15433
CNNscore: 0.30002
CNNaffinity: 7.48199
CNNvariance: 0.67574
Affinity: 4.91153 -0.06648 (kcal/mol)
RMSD: 0.29442
CNNscore: 0.17850
CNNaffinity: 7.57060
CNNvariance: 0.18979
Affinity: 3.81685 -0.71248 (kcal/mol)
RMSD: 0.06638
CNNscore: 0.10292
CNNaffinity: 7.40983
CNNvariance: 0.21940
Affinity: 5.19123 -1.65243 (kcal/mol)
RMSD: 0.67882
CNNscore: 0.56503
CNNaffinity: 8.01951
CNNvariance: 0.74790
Affinity: 3.04622 0.24981 (kcal/mol)
RMSD: 0.32951
CNNscore: 0.44788
CNNaffinity: 7.79804
CNNvariance: 0.65795
Affinity: 5.67593 -2.30507 (kcal/mol)
RMSD: 0.20291
CNNscore: 0.41337
CNNaffinity: 7.88850
CNNvariance: 0.19241
Affinity: 3.76098 -2.38548 (kcal/mol)
RMSD: 0.12702
CNNscore: 0.06836
CNNaffinity: 7.26938
CNNvariance: 0.21528
Affinity: 3.39981 -1.62878 (kcal/mol)
RMSD: 0.28075
CNNscore: 0.05067
CNNaffinity: 7.06565
CNNvariance: 0.28753
Affinity: 5.10709 -0.76718 (kcal/mol)
RMSD: 0.19136
CNNscore: 0.28424
CNNaffinity: 7.48877
CNNvariance: 0.54235
Affinity: 4.46925 -0.74674 (kcal/mol)
RMSD: 0.98400
CNNscore: 0.14920
CNNaffinity: 7.33165
CNNvariance: 0.66776
Affinity: 2.04725 -1.80269 (kcal/mol)
RMSD: 0.80565
CNNscore: 0.18398
CNNaffinity: 7.64864
CNNvariance: 0.43049
Affinity: 4.23508 -2.53209 (kcal/mol)
RMSD: 0.36806
CNNscore: 0.36531
CNNaffinity: 7.56819
CNNvariance: 0.60570
Affinity: 5.44198 -1.38624 (kcal/mol)
RMSD: 0.13374
CNNscore: 0.18351
CNNaffinity: 7.42353
CNNvariance: 0.18012
Affinity: 5.96900 -0.24676 (kcal/mol)
RMSD: 0.19906
CNNscore: 0.15656
CNNaffinity: 6.69105
CNNvariance: 0.03005
Affinity: 5.32178 1.44536 (kcal/mol)
RMSD: 0.22222
CNNscore: 0.11352
CNNaffinity: 7.29612
CNNvariance: 0.18698
Affinity: 6.11795 -1.41001 (kcal/mol)
RMSD: 0.21551
CNNscore: 0.24409
CNNaffinity: 7.36383
CNNvariance: 0.23513
Affinity: 5.31928 1.54306 (kcal/mol)
RMSD: 0.20691
CNNscore: 0.29070
CNNaffinity: 7.34406
CNNvariance: 0.45304
Affinity: 6.22313 -0.62553 (kcal/mol)
RMSD: 0.15859
CNNscore: 0.11991
CNNaffinity: 7.25967
CNNvariance: 0.36662
Affinity: 3.01656 -0.62940 (kcal/mol)
RMSD: 0.39024
CNNscore: 0.13488
CNNaffinity: 7.15718
CNNvariance: 0.10499
Affinity: 6.58981 2.64457 (kcal/mol)
RMSD: 0.25838
CNNscore: 0.20192
CNNaffinity: 7.47832
CNNvariance: 0.15747
Affinity: 7.22339 4.42293 (kcal/mol)
RMSD: 0.25110
CNNscore: 0.18861
CNNaffinity: 7.51264
CNNvariance: 0.22233
Affinity: 5.33184 -1.06670 (kcal/mol)
RMSD: 0.21384
CNNscore: 0.20441
CNNaffinity: 7.77242
CNNvariance: 0.45518
Affinity: 7.62209 0.71010 (kcal/mol)
RMSD: 0.44579
CNNscore: 0.46768
CNNaffinity: 8.21212
CNNvariance: 0.81952
Affinity: 6.41883 -0.91638 (kcal/mol)
RMSD: 0.20117
CNNscore: 0.28011
CNNaffinity: 7.07851
CNNvariance: 0.22859
Affinity: 6.99984 0.05997 (kcal/mol)
RMSD: 0.11664
CNNscore: 0.39983
CNNaffinity: 7.62404
CNNvariance: 0.31290
Affinity: 5.47973 0.05815 (kcal/mol)
RMSD: 0.13192
CNNscore: 0.20642
CNNaffinity: 7.20169
CNNvariance: 0.13487
Affinity: 5.41293 0.58190 (kcal/mol)
RMSD: 0.27180
CNNscore: 0.25831
CNNaffinity: 7.58506
CNNvariance: 0.14365
Affinity: 7.55615 1.98193 (kcal/mol)
RMSD: 0.41708
CNNscore: 0.11533
CNNaffinity: 7.25042
CNNvariance: 0.11536
Affinity: 5.09547 -0.34588 (kcal/mol)
RMSD: 0.19491
CNNscore: 0.20829
CNNaffinity: 7.43469
CNNvariance: 0.22909
Affinity: 6.89905 -0.07916 (kcal/mol)
RMSD: 0.11889
CNNscore: 0.26849
CNNaffinity: 7.56069
CNNvariance: 0.08511
Affinity: 3.72852 5.21657 (kcal/mol)
RMSD: 0.23774
CNNscore: 0.08725
CNNaffinity: 7.31067
CNNvariance: 0.11357
Affinity: 4.52627 0.40014 (kcal/mol)
RMSD: 0.11016
CNNscore: 0.06142
CNNaffinity: 7.04779
CNNvariance: 0.59107
Affinity: 8.00680 -1.47233 (kcal/mol)
RMSD: 0.28818
CNNscore: 0.35556
CNNaffinity: 7.15888
CNNvariance: 0.44502
Affinity: 5.77661 -0.63584 (kcal/mol)
RMSD: 0.31389
CNNscore: 0.18836
CNNaffinity: 7.34028
CNNvariance: 0.18388
Affinity: -0.57760 -0.51886 (kcal/mol)
RMSD: 0.58898
CNNscore: 0.28636
CNNaffinity: 7.18173
CNNvariance: 0.42582
Affinity: 5.94926 -1.06678 (kcal/mol)
RMSD: 1.11848
CNNscore: 0.30732
CNNaffinity: 6.42264
CNNvariance: 0.73701
Affinity: 7.26665 -3.20742 (kcal/mol)
RMSD: 0.16189
CNNscore: 0.31071
CNNaffinity: 7.93612
CNNvariance: 0.47788
Affinity: 5.05074 0.26192 (kcal/mol)
RMSD: 0.19097
CNNscore: 0.46480
CNNaffinity: 8.18236
CNNvariance: 0.38751
Affinity: 6.62505 -0.59789 (kcal/mol)
RMSD: 0.16816
CNNscore: 0.29396
CNNaffinity: 7.67189
CNNvariance: 0.57546
Affinity: 6.26048 -1.29869 (kcal/mol)
RMSD: 0.18011
CNNscore: 0.14902
CNNaffinity: 7.78565
CNNvariance: 0.35120
Affinity: 8.51398 0.69563 (kcal/mol)
RMSD: 0.20522
CNNscore: 0.40541
CNNaffinity: 7.70596
CNNvariance: 0.16211
Affinity: 4.16854 0.13015 (kcal/mol)
RMSD: 0.19907
CNNscore: 0.28371
CNNaffinity: 7.60733
CNNvariance: 1.43814
Affinity: 6.51914 -0.97833 (kcal/mol)
RMSD: 0.15122
CNNscore: 0.19233
CNNaffinity: 7.34283
CNNvariance: 0.16221
Affinity: 7.82773 0.90879 (kcal/mol)
RMSD: 0.26518
CNNscore: 0.16504
CNNaffinity: 7.30118
CNNvariance: 0.12807
Affinity: 5.52804 -2.15691 (kcal/mol)
RMSD: 0.18008
CNNscore: 0.44297
CNNaffinity: 7.96293
CNNvariance: 0.52001
Affinity: 5.89938 -1.03560 (kcal/mol)
RMSD: 0.55172
CNNscore: 0.34836
CNNaffinity: 7.78687
CNNvariance: 0.72682
Affinity: 5.66484 -1.00320 (kcal/mol)
RMSD: 0.10650
CNNscore: 0.18450
CNNaffinity: 7.52252
CNNvariance: 0.11238
Affinity: 7.41244 -1.91383 (kcal/mol)
RMSD: 0.19345
CNNscore: 0.47826
CNNaffinity: 7.62673
CNNvariance: 0.55560
Affinity: 9.98551 6.21004 (kcal/mol)
RMSD: 0.21029
CNNscore: 0.37699
CNNaffinity: 7.55418
CNNvariance: 0.17075
Affinity: 6.72341 0.02432 (kcal/mol)
RMSD: 0.42867
CNNscore: 0.25150
CNNaffinity: 6.94248
CNNvariance: 0.28923
Affinity: 6.04181 1.25586 (kcal/mol)
RMSD: 0.25068
CNNscore: 0.33264
CNNaffinity: 7.89999
CNNvariance: 0.52088
Affinity: 6.78638 -0.15985 (kcal/mol)
RMSD: 0.13783
CNNscore: 0.18756
CNNaffinity: 7.40092
CNNvariance: 0.13034
Affinity: 6.14539 -0.26411 (kcal/mol)
RMSD: 0.21892
CNNscore: 0.33931
CNNaffinity: 7.74732
CNNvariance: 0.23270
Affinity: 7.94778 0.49460 (kcal/mol)
RMSD: 0.40658
CNNscore: 0.27499
CNNaffinity: 7.76342
CNNvariance: 0.25982
Affinity: 5.24862 -0.32405 (kcal/mol)
RMSD: 0.17982
CNNscore: 0.09505
CNNaffinity: 7.47555
CNNvariance: 0.17835
Affinity: 8.82442 -1.41842 (kcal/mol)
RMSD: 0.21265
CNNscore: 0.43831
CNNaffinity: 7.74492
CNNvariance: 0.89107
Affinity: 5.59867 0.35792 (kcal/mol)
RMSD: 1.00137
CNNscore: 0.47334
CNNaffinity: 7.68989
CNNvariance: 0.41705
Affinity: 1.63592 -0.21989 (kcal/mol)
RMSD: 1.93836
CNNscore: 0.35754
CNNaffinity: 7.06373
CNNvariance: 0.13101
Affinity: 7.00145 2.10081 (kcal/mol)
RMSD: 0.12164
CNNscore: 0.09636
CNNaffinity: 7.37462
CNNvariance: 0.19946
Affinity: 5.92397 -0.40971 (kcal/mol)
RMSD: 1.09223
CNNscore: 0.25049
CNNaffinity: 6.94398
CNNvariance: 0.87312
Affinity: 6.48192 -2.35292 (kcal/mol)
RMSD: 0.13038
CNNscore: 0.47628
CNNaffinity: 7.95238
CNNvariance: 0.46648
Affinity: 5.93138 -0.64862 (kcal/mol)
RMSD: 0.16435
CNNscore: 0.04598
CNNaffinity: 7.28603
CNNvariance: 0.40020
Affinity: 7.07983 -1.26486 (kcal/mol)
RMSD: 0.79781
CNNscore: 0.16396
CNNaffinity: 7.28218
CNNvariance: 0.36346
Affinity: 7.43065 0.16814 (kcal/mol)
RMSD: 0.21721
CNNscore: 0.22576
CNNaffinity: 7.60256
CNNvariance: 0.11387
Affinity: 6.44982 -0.95866 (kcal/mol)
RMSD: 0.38269
CNNscore: 0.30616
CNNaffinity: 6.95255
CNNvariance: 1.03878
Affinity: -2.05955 -0.54860 (kcal/mol)
RMSD: 2.28331
CNNscore: 0.31213
CNNaffinity: 6.28155
CNNvariance: 0.41865
Affinity: 9.71193 1.48784 (kcal/mol)
RMSD: 0.27165
CNNscore: 0.16330
CNNaffinity: 7.24638
CNNvariance: 0.26608
Affinity: 7.85825 -1.05102 (kcal/mol)
RMSD: 0.40653
CNNscore: 0.20352
CNNaffinity: 7.50065
CNNvariance: 0.37503
Affinity: 7.40760 0.58640 (kcal/mol)
RMSD: 0.15639
CNNscore: 0.15408
CNNaffinity: 7.48596
CNNvariance: 0.36101
Affinity: 8.28067 -1.27813 (kcal/mol)
RMSD: 0.08459
CNNscore: 0.39376
CNNaffinity: 7.96965
CNNvariance: 0.43459
Affinity: 9.83268 -0.63076 (kcal/mol)
RMSD: 0.34732
CNNscore: 0.25771
CNNaffinity: 7.81733
CNNvariance: 0.12442
Affinity: 6.97095 -1.13368 (kcal/mol)
RMSD: 1.45148
CNNscore: 0.10799
CNNaffinity: 7.22008
CNNvariance: 0.28320
Affinity: 7.23795 0.14258 (kcal/mol)
RMSD: 0.23429
CNNscore: 0.40514
CNNaffinity: 7.87891
CNNvariance: 1.54071
Affinity: 8.54613 0.11563 (kcal/mol)
RMSD: 0.80844
CNNscore: 0.29713
CNNaffinity: 7.53094
CNNvariance: 0.42486
Affinity: 8.02957 1.20949 (kcal/mol)
RMSD: 0.37955
CNNscore: 0.18883
CNNaffinity: 7.55787
CNNvariance: 0.33722
Affinity: 10.39096 0.38866 (kcal/mol)
RMSD: 0.58032
CNNscore: 0.30604
CNNaffinity: 7.58923
CNNvariance: 0.43184
Affinity: 7.44544 -0.64827 (kcal/mol)
RMSD: 0.55202
CNNscore: 0.28807
CNNaffinity: 7.55959
CNNvariance: 1.49905
Affinity: 5.49987 -1.06142 (kcal/mol)
RMSD: 0.20622
CNNscore: 0.25472
CNNaffinity: 6.89020
CNNvariance: 0.72087
Affinity: -1.33999 -0.48610 (kcal/mol)
RMSD: 1.93215
CNNscore: 0.31984
CNNaffinity: 6.49728
CNNvariance: 0.88133
Affinity: 6.24102 -0.06737 (kcal/mol)
RMSD: 0.15826
CNNscore: 0.39604
CNNaffinity: 7.49653
CNNvariance: 0.95666
Affinity: 8.35378 2.57196 (kcal/mol)
RMSD: 0.18775
CNNscore: 0.07247
CNNaffinity: 7.33167
CNNvariance: 0.43591
Affinity: 9.11855 0.61585 (kcal/mol)
RMSD: 0.15158
CNNscore: 0.26220
CNNaffinity: 7.29112
CNNvariance: 0.08909
Affinity: 8.44928 -1.13790 (kcal/mol)
RMSD: 0.16109
CNNscore: 0.38667
CNNaffinity: 7.70227
CNNvariance: 0.44901
Affinity: 11.01611 0.00446 (kcal/mol)
RMSD: 0.40794
CNNscore: 0.44588
CNNaffinity: 7.61489
CNNvariance: 2.14119
Affinity: 6.10831 -0.48559 (kcal/mol)
RMSD: 1.50920
CNNscore: 0.38102
CNNaffinity: 7.25889
CNNvariance: 0.51198
Affinity: 10.45841 -1.03432 (kcal/mol)
RMSD: 0.11940
CNNscore: 0.26228
CNNaffinity: 7.61554
CNNvariance: 0.15167
Affinity: 1.31964 -0.82603 (kcal/mol)
RMSD: 1.90150
CNNscore: 0.56543
CNNaffinity: 7.07048
CNNvariance: 0.33446
Affinity: 8.87351 0.90496 (kcal/mol)
RMSD: 0.13918
CNNscore: 0.29175
CNNaffinity: 7.66827
CNNvariance: 0.15886
Affinity: 9.78569 -1.95052 (kcal/mol)
RMSD: 0.74436
CNNscore: 0.38580
CNNaffinity: 7.93808
CNNvariance: 0.41122
Affinity: 10.56977 -1.04007 (kcal/mol)
RMSD: 1.12317
CNNscore: 0.28955
CNNaffinity: 7.29395
CNNvariance: 0.32169
Affinity: 11.73677 -0.26569 (kcal/mol)
RMSD: 0.68752
CNNscore: 0.48717
CNNaffinity: 8.01853
CNNvariance: 0.19640
Affinity: 12.37902 -1.22105 (kcal/mol)
RMSD: 0.35554
CNNscore: 0.47744
CNNaffinity: 8.01317
CNNvariance: 0.68585
Affinity: 11.38974 0.08313 (kcal/mol)
RMSD: 0.16310
CNNscore: 0.40953
CNNaffinity: 7.90877
CNNvariance: 0.71652
Affinity: 10.60369 -0.10345 (kcal/mol)
RMSD: 0.22788
CNNscore: 0.28633
CNNaffinity: 7.49764
CNNvariance: 0.10777
Affinity: 9.23495 2.15640 (kcal/mol)
RMSD: 0.15424
CNNscore: 0.37593
CNNaffinity: 7.72100
CNNvariance: 1.90460
Affinity: 7.47061 -0.44624 (kcal/mol)
RMSD: 0.14293
CNNscore: 0.33512
CNNaffinity: 7.33484
CNNvariance: 0.68610
Affinity: 15.20939 3.41524 (kcal/mol)
RMSD: 0.16506
CNNscore: 0.29173
CNNaffinity: 7.89948
CNNvariance: 0.41588
Affinity: 14.00523 -1.19226 (kcal/mol)
RMSD: 0.15965
CNNscore: 0.38016
CNNaffinity: 8.01146
CNNvariance: 0.64954
Affinity: 9.41203 -0.72378 (kcal/mol)
RMSD: 0.07454
CNNscore: 0.25691
CNNaffinity: 7.26137
CNNvariance: 1.12603
Affinity: 11.98184 0.49403 (kcal/mol)
RMSD: 0.16148
CNNscore: 0.22702
CNNaffinity: 7.99019
CNNvariance: 0.45539
Affinity: 7.85199 -0.81895 (kcal/mol)
RMSD: 0.55424
CNNscore: 0.23617
CNNaffinity: 7.09167
CNNvariance: 0.35494
Affinity: 8.36232 -1.79918 (kcal/mol)
RMSD: 0.43772
CNNscore: 0.13946
CNNaffinity: 7.51506
CNNvariance: 0.15251
Affinity: 6.55964 -1.00447 (kcal/mol)
RMSD: 0.92395
CNNscore: 0.46893
CNNaffinity: 7.81790
CNNvariance: 0.48042
Affinity: 9.75320 2.83398 (kcal/mol)
RMSD: 0.69193
CNNscore: 0.31242
CNNaffinity: 7.59371
CNNvariance: 0.29048
Affinity: 8.80496 -1.02408 (kcal/mol)
RMSD: 0.18759
CNNscore: 0.19873
CNNaffinity: 6.69441
CNNvariance: 0.29656
Affinity: 10.87033 -0.11658 (kcal/mol)
RMSD: 0.32442
CNNscore: 0.07064
CNNaffinity: 7.25392
CNNvariance: 0.26008
Affinity: 16.16102 -1.00619 (kcal/mol)
RMSD: 0.21036
CNNscore: 0.21087
CNNaffinity: 8.17338
CNNvariance: 0.14897
Affinity: 11.99878 -2.21900 (kcal/mol)
RMSD: 0.19367
CNNscore: 0.38299
CNNaffinity: 7.89677
CNNvariance: 1.19351
Affinity: 10.05201 -0.97314 (kcal/mol)
RMSD: 0.23526
CNNscore: 0.25246
CNNaffinity: 7.72727
CNNvariance: 0.55241
Affinity: 11.26044 -1.13922 (kcal/mol)
RMSD: 0.19973
CNNscore: 0.16158
CNNaffinity: 7.34502
CNNvariance: 0.12706
Affinity: 0.61076 -0.33353 (kcal/mol)
RMSD: 2.84095
CNNscore: 0.54356
CNNaffinity: 6.67206
CNNvariance: 0.43864
Affinity: 7.84366 0.76079 (kcal/mol)
RMSD: 1.75001
CNNscore: 0.11545
CNNaffinity: 7.38721
CNNvariance: 0.15306
Affinity: 9.14185 -0.81424 (kcal/mol)
RMSD: 0.10244
CNNscore: 0.58527
CNNaffinity: 8.25173
CNNvariance: 1.59486
Affinity: 11.05315 -1.24876 (kcal/mol)
RMSD: 0.33075
CNNscore: 0.04786
CNNaffinity: 7.18894
CNNvariance: 0.11144
Affinity: 9.51059 -1.06873 (kcal/mol)
RMSD: 0.16119
CNNscore: 0.31815
CNNaffinity: 6.91224
CNNvariance: 0.49582
Affinity: 9.01496 -2.85478 (kcal/mol)
RMSD: 1.53375
CNNscore: 0.19690
CNNaffinity: 7.66995
CNNvariance: 0.42509
Affinity: 10.29252 -1.74107 (kcal/mol)
RMSD: 1.69407
CNNscore: 0.31636
CNNaffinity: 7.54306
CNNvariance: 0.42935
Affinity: 14.59426 -1.71353 (kcal/mol)
RMSD: 0.34245
CNNscore: 0.07115
CNNaffinity: 7.60565
CNNvariance: 0.20465
Affinity: 15.02647 -3.20386 (kcal/mol)
RMSD: 0.13749
CNNscore: 0.16546
CNNaffinity: 7.48324
CNNvariance: 0.11124
Affinity: 14.33425 -0.73282 (kcal/mol)
RMSD: 0.32325
CNNscore: 0.23704
CNNaffinity: 7.64627
CNNvariance: 0.44642
Affinity: 10.74973 -1.70260 (kcal/mol)
RMSD: 0.23842
CNNscore: 0.15055
CNNaffinity: 7.07909
CNNvariance: 0.78386
Affinity: 10.15740 -1.87328 (kcal/mol)
RMSD: 0.38592
CNNscore: 0.29576
CNNaffinity: 7.76739
CNNvariance: 1.12855
Affinity: 6.77273 -0.89292 (kcal/mol)
RMSD: 0.59947
CNNscore: 0.19865
CNNaffinity: 7.18884
CNNvariance: 0.18743
Affinity: 13.86529 -1.63340 (kcal/mol)
RMSD: 0.20246
CNNscore: 0.47600
CNNaffinity: 8.14911
CNNvariance: 0.83498
Affinity: 13.04817 -0.36800 (kcal/mol)
RMSD: 0.24772
CNNscore: 0.29163
CNNaffinity: 7.59849
CNNvariance: 0.54253
Affinity: 14.92050 -1.77732 (kcal/mol)
RMSD: 0.33287
CNNscore: 0.10460
CNNaffinity: 7.57730
CNNvariance: 0.30095
Affinity: 10.31842 -0.64985 (kcal/mol)
RMSD: 0.42263
CNNscore: 0.10668
CNNaffinity: 7.63218
CNNvariance: 1.20693
Affinity: 5.12450 -0.88759 (kcal/mol)
RMSD: 1.89860
CNNscore: 0.48735
CNNaffinity: 6.74473
CNNvariance: 0.58804
Affinity: 11.78547 -1.21270 (kcal/mol)
RMSD: 0.13978
CNNscore: 0.47618
CNNaffinity: 7.74823
CNNvariance: 0.87041
Affinity: 11.94124 -1.59554 (kcal/mol)
RMSD: 0.09318
CNNscore: 0.40184
CNNaffinity: 7.90295
CNNvariance: 0.35673
Affinity: 13.53569 0.26615 (kcal/mol)
RMSD: 0.22192
CNNscore: 0.41237
CNNaffinity: 7.73919
CNNvariance: 0.31814
Affinity: 13.77286 0.42229 (kcal/mol)
RMSD: 0.33031
CNNscore: 0.27678
CNNaffinity: 7.65105
CNNvariance: 0.39332
Affinity: 12.11834 -1.31523 (kcal/mol)
RMSD: 0.97327
CNNscore: 0.04334
CNNaffinity: 6.91341
CNNvariance: 0.04847
Affinity: 13.39227 0.71338 (kcal/mol)
RMSD: 0.34491
CNNscore: 0.31711
CNNaffinity: 7.31465
CNNvariance: 0.61691
Affinity: 9.69012 -1.24891 (kcal/mol)
RMSD: 0.58243
CNNscore: 0.35593
CNNaffinity: 7.79553
CNNvariance: 0.41607
Affinity: 6.64078 -0.93336 (kcal/mol)
RMSD: 1.89519
CNNscore: 0.43171
CNNaffinity: 7.07216
CNNvariance: 0.46701
Affinity: 14.43615 -1.01298 (kcal/mol)
RMSD: 0.16203
CNNscore: 0.39765
CNNaffinity: 7.91352
CNNvariance: 0.93607
Affinity: 12.12180 -1.48959 (kcal/mol)
RMSD: 0.57687
CNNscore: 0.30328
CNNaffinity: 7.45961
CNNvariance: 0.54316
Affinity: 13.53675 0.76924 (kcal/mol)
RMSD: 0.84946
CNNscore: 0.36518
CNNaffinity: 7.47913
CNNvariance: 0.35209
Affinity: 12.91976 1.22459 (kcal/mol)
RMSD: 0.38371
CNNscore: 0.07330
CNNaffinity: 7.56524
CNNvariance: 0.36981
Affinity: 12.18681 -1.70441 (kcal/mol)
RMSD: 0.12020
CNNscore: 0.31460
CNNaffinity: 7.68508
CNNvariance: 0.45513
Affinity: 14.62400 -1.42222 (kcal/mol)
RMSD: 0.70516
CNNscore: 0.43838
CNNaffinity: 7.94416
CNNvariance: 0.52795
Affinity: 14.24045 -1.74765 (kcal/mol)
RMSD: 0.18061
CNNscore: 0.20746
CNNaffinity: 7.36441
CNNvariance: 0.59945
Affinity: 13.52623 -1.99147 (kcal/mol)
RMSD: 0.24873
CNNscore: 0.15062
CNNaffinity: 7.33882
CNNvariance: 0.52412
Affinity: 14.95494 -2.10597 (kcal/mol)
RMSD: 0.16366
CNNscore: 0.16463
CNNaffinity: 7.19069
CNNvariance: 0.15574
Affinity: 13.62823 -1.71168 (kcal/mol)
RMSD: 0.27091
CNNscore: 0.22607
CNNaffinity: 7.31361
CNNvariance: 0.63215
Affinity: 15.30974 -0.30478 (kcal/mol)
RMSD: 1.23698
CNNscore: 0.10589
CNNaffinity: 7.64053
CNNvariance: 0.17445
Affinity: 13.99472 -1.98804 (kcal/mol)
RMSD: 0.32538
CNNscore: 0.19272
CNNaffinity: 7.37921
CNNvariance: 0.63011
Affinity: 15.44396 0.00345 (kcal/mol)
RMSD: 0.42315
CNNscore: 0.24559
CNNaffinity: 7.68023
CNNvariance: 0.70563
Affinity: 12.24938 -0.08654 (kcal/mol)
RMSD: 0.95766
CNNscore: 0.06088
CNNaffinity: 7.21387
CNNvariance: 0.21899
Affinity: 0.02909 0.06869 (kcal/mol)
RMSD: 2.49398
CNNscore: 0.45179
CNNaffinity: 7.65297
CNNvariance: 0.46242
Affinity: 13.45993 1.21218 (kcal/mol)
RMSD: 0.15493
CNNscore: 0.16194
CNNaffinity: 7.16350
CNNvariance: 0.51017
Affinity: 12.70368 0.32374 (kcal/mol)
RMSD: 0.15993
CNNscore: 0.29832
CNNaffinity: 7.67485
CNNvariance: 1.41969
Affinity: 2.25970 -1.07158 (kcal/mol)
RMSD: 2.65620
CNNscore: 0.56823
CNNaffinity: 7.28575
CNNvariance: 0.33481
Affinity: 11.36205 -0.70459 (kcal/mol)
RMSD: 0.06420
CNNscore: 0.47372
CNNaffinity: 7.75417
CNNvariance: 1.72921
Affinity: 16.82206 -0.93669 (kcal/mol)
RMSD: 0.84633
CNNscore: 0.41128
CNNaffinity: 7.79787
CNNvariance: 0.50013
Affinity: 14.77679 -1.94758 (kcal/mol)
RMSD: 0.11175
CNNscore: 0.26087
CNNaffinity: 7.19991
CNNvariance: 0.81778
Affinity: 13.52121 -1.44564 (kcal/mol)
RMSD: 0.23204
CNNscore: 0.32762
CNNaffinity: 7.41931
CNNvariance: 0.82620
Affinity: 14.15446 -1.92686 (kcal/mol)
RMSD: 0.68799
CNNscore: 0.16062
CNNaffinity: 7.63995
CNNvariance: 0.29287
Affinity: 16.71346 -0.35576 (kcal/mol)
RMSD: 0.12605
CNNscore: 0.60404
CNNaffinity: 8.04486
CNNvariance: 0.75663
Affinity: 12.36725 1.62905 (kcal/mol)
RMSD: 0.24588
CNNscore: 0.18553
CNNaffinity: 6.41367
CNNvariance: 0.33457
Affinity: 7.87985 0.51290 (kcal/mol)
RMSD: 1.62297
CNNscore: 0.34425
CNNaffinity: 6.78239
CNNvariance: 0.32595
Affinity: 11.48329 -0.53139 (kcal/mol)
RMSD: 0.75930
CNNscore: 0.21487
CNNaffinity: 6.58164
CNNvariance: 0.80115
Affinity: 17.51661 1.32673 (kcal/mol)
RMSD: 0.12056
CNNscore: 0.20972
CNNaffinity: 7.20064
CNNvariance: 0.51301
Affinity: 10.68447 -1.48731 (kcal/mol)
RMSD: 0.81802
CNNscore: 0.28787
CNNaffinity: 7.43871
CNNvariance: 0.72990
Affinity: 15.46414 1.07508 (kcal/mol)
RMSD: 1.47605
CNNscore: 0.41412
CNNaffinity: 7.53659
CNNvariance: 1.11088
Affinity: 18.28492 -3.50242 (kcal/mol)
RMSD: 0.11803
CNNscore: 0.20184
CNNaffinity: 7.46260
CNNvariance: 0.19866
Affinity: 12.84370 -0.16490 (kcal/mol)
RMSD: 0.58122
CNNscore: 0.47662
CNNaffinity: 7.63586
CNNvariance: 1.63136
Affinity: 14.73333 -1.76758 (kcal/mol)
RMSD: 0.67168
CNNscore: 0.17762
CNNaffinity: 7.44641
CNNvariance: 0.47703
Affinity: 18.76616 2.04279 (kcal/mol)
RMSD: 0.37633
CNNscore: 0.45621
CNNaffinity: 7.72660
CNNvariance: 0.82139
Affinity: 16.88579 -0.79721 (kcal/mol)
RMSD: 0.54971
CNNscore: 0.21984
CNNaffinity: 7.56305
CNNvariance: 0.94378
Affinity: 18.29575 0.34864 (kcal/mol)
RMSD: 0.26203
CNNscore: 0.27654
CNNaffinity: 7.35022
CNNvariance: 0.83784
Affinity: 25.02318 2.47621 (kcal/mol)
RMSD: 0.20977
CNNscore: 0.16151
CNNaffinity: 7.73830
CNNvariance: 0.27529
Affinity: 17.54648 -0.97630 (kcal/mol)
RMSD: 0.11590
CNNscore: 0.49313
CNNaffinity: 7.71544
CNNvariance: 1.85603
Affinity: 18.87146 -1.81953 (kcal/mol)
RMSD: 0.70432
CNNscore: 0.17098
CNNaffinity: 7.72027
CNNvariance: 0.32825
Affinity: 21.94052 0.33009 (kcal/mol)
RMSD: 0.21985
CNNscore: 0.60105
CNNaffinity: 7.98295
CNNvariance: 0.29352
Affinity: 18.15722 -1.56540 (kcal/mol)
RMSD: 0.19006
CNNscore: 0.15940
CNNaffinity: 7.28271
CNNvariance: 0.18817
Affinity: 6.52811 -0.46744 (kcal/mol)
RMSD: 2.04243
CNNscore: 0.13658
CNNaffinity: 7.48472
CNNvariance: 0.20984
Affinity: 11.91002 -0.81367 (kcal/mol)
RMSD: 0.99984
CNNscore: 0.38114
CNNaffinity: 7.63425
CNNvariance: 1.92209
Affinity: 16.34028 -1.71936 (kcal/mol)
RMSD: 0.60205
CNNscore: 0.19161
CNNaffinity: 7.33674
CNNvariance: 0.60570
Affinity: 16.89801 1.89965 (kcal/mol)
RMSD: 0.21096
CNNscore: 0.27579
CNNaffinity: 7.43982
CNNvariance: 0.36054
Affinity: 20.08518 -0.23737 (kcal/mol)
RMSD: 0.14113
CNNscore: 0.30889
CNNaffinity: 7.49567
CNNvariance: 0.36928
Affinity: 12.70146 -0.30084 (kcal/mol)
RMSD: 1.57764
CNNscore: 0.10116
CNNaffinity: 6.92698
CNNvariance: 0.39470
Affinity: 18.66435 0.30307 (kcal/mol)
RMSD: 0.31429
CNNscore: 0.13646
CNNaffinity: 6.92383
CNNvariance: 0.31421
Affinity: 18.01520 -1.94919 (kcal/mol)
RMSD: 0.93497
CNNscore: 0.39937
CNNaffinity: 7.62459
CNNvariance: 0.34731
Affinity: 19.16951 -0.18865 (kcal/mol)
RMSD: 0.10624
CNNscore: 0.13287
CNNaffinity: 7.17966
CNNvariance: 0.46101
Affinity: 21.94008 -0.95757 (kcal/mol)
RMSD: 0.09383
CNNscore: 0.44992
CNNaffinity: 7.25202
CNNvariance: 0.51240
Affinity: 22.31860 0.04834 (kcal/mol)
RMSD: 0.11800
CNNscore: 0.38875
CNNaffinity: 7.51456
CNNvariance: 0.64122
Affinity: 6.84760 -0.46842 (kcal/mol)
RMSD: 3.36601
CNNscore: 0.14352
CNNaffinity: 7.41896
CNNvariance: 0.55368
Affinity: 21.38613 0.54447 (kcal/mol)
RMSD: 0.27040
CNNscore: 0.26030
CNNaffinity: 7.41307
CNNvariance: 0.55028
Affinity: 19.20714 -1.43387 (kcal/mol)
RMSD: 0.19854
CNNscore: 0.14478
CNNaffinity: 7.17102
CNNvariance: 0.28042
Affinity: 19.36029 -2.39717 (kcal/mol)
RMSD: 1.01171
CNNscore: 0.15649
CNNaffinity: 7.45805
CNNvariance: 0.56510
Affinity: 15.31450 2.14884 (kcal/mol)
RMSD: 0.64366
CNNscore: 0.09008
CNNaffinity: 7.14863
CNNvariance: 0.38040
Affinity: 6.88341 -0.02509 (kcal/mol)
RMSD: 4.17772
CNNscore: 0.24902
CNNaffinity: 6.79340
CNNvariance: 0.18427
Affinity: 14.37022 0.93096 (kcal/mol)
RMSD: 2.10675
CNNscore: 0.05587
CNNaffinity: 7.20062
CNNvariance: 0.12494
Affinity: 19.72075 -3.75152 (kcal/mol)
RMSD: 0.39211
CNNscore: 0.49246
CNNaffinity: 8.34470
CNNvariance: 0.55940
Affinity: 1.07489 -2.66495 (kcal/mol)
RMSD: 3.52771
CNNscore: 0.13741
CNNaffinity: 7.44720
CNNvariance: 0.48838
Affinity: 22.27999 -3.10929 (kcal/mol)
RMSD: 0.29493
CNNscore: 0.35984
CNNaffinity: 8.01283
CNNvariance: 0.50808
Affinity: 22.64667 1.22919 (kcal/mol)
RMSD: 0.12349
CNNscore: 0.40339
CNNaffinity: 7.09059
CNNvariance: 0.72309
Affinity: 25.44596 -0.35296 (kcal/mol)
RMSD: 0.72859
CNNscore: 0.16778
CNNaffinity: 7.26908
CNNvariance: 0.31162
Affinity: 25.52406 -0.90391 (kcal/mol)
RMSD: 0.16810
CNNscore: 0.26856
CNNaffinity: 7.64746
CNNvariance: 1.01735
Affinity: 25.16447 -1.84332 (kcal/mol)
RMSD: 0.12786
CNNscore: 0.15510
CNNaffinity: 6.95679
CNNvariance: 0.31144
Affinity: 15.28719 -0.06483 (kcal/mol)
RMSD: 2.81875
CNNscore: 0.16211
CNNaffinity: 7.67974
CNNvariance: 0.56799
Affinity: 4.94649 1.06000 (kcal/mol)
RMSD: 3.67873
CNNscore: 0.16820
CNNaffinity: 7.51068
CNNvariance: 0.35592
Affinity: 24.58359 -0.62966 (kcal/mol)
RMSD: 1.67650
CNNscore: 0.07546
CNNaffinity: 6.71146
CNNvariance: 0.71076
Affinity: 18.84138 0.24484 (kcal/mol)
RMSD: 1.31954
CNNscore: 0.22532
CNNaffinity: 6.51322
CNNvariance: 0.37263
Affinity: 25.85871 -0.10182 (kcal/mol)
RMSD: 0.11206
CNNscore: 0.57685
CNNaffinity: 7.98715
CNNvariance: 2.42195
Affinity: 30.26468 -2.24879 (kcal/mol)
RMSD: 0.20485
CNNscore: 0.16436
CNNaffinity: 7.52160
CNNvariance: 0.71539
Affinity: 23.70237 0.59015 (kcal/mol)
RMSD: 1.04689
CNNscore: 0.51627
CNNaffinity: 7.62860
CNNvariance: 1.32364
Affinity: 24.26942 1.18650 (kcal/mol)
RMSD: 0.87610
CNNscore: 0.42121
CNNaffinity: 7.12277
CNNvariance: 1.31327
Affinity: 28.61276 -2.51876 (kcal/mol)
RMSD: 0.44694
CNNscore: 0.21212
CNNaffinity: 7.91584
CNNvariance: 0.54460
Affinity: 24.55962 -0.98447 (kcal/mol)
RMSD: 0.16404
CNNscore: 0.20245
CNNaffinity: 6.98098
CNNvariance: 0.39022
Affinity: 29.54314 -2.74884 (kcal/mol)
RMSD: 1.26490
CNNscore: 0.29899
CNNaffinity: 7.72887
CNNvariance: 0.31495
Affinity: 21.36326 1.05634 (kcal/mol)
RMSD: 0.56395
CNNscore: 0.09573
CNNaffinity: 7.32673
CNNvariance: 0.90037
Affinity: 23.06244 -2.59292 (kcal/mol)
RMSD: 1.07548
CNNscore: 0.19688
CNNaffinity: 7.35938
CNNvariance: 0.66579
Affinity: 27.76864 -2.82496 (kcal/mol)
RMSD: 0.34068
CNNscore: 0.39174
CNNaffinity: 8.16902
CNNvariance: 0.48965
Affinity: 30.77102 -1.20416 (kcal/mol)
RMSD: 0.24197
CNNscore: 0.32591
CNNaffinity: 7.94227
CNNvariance: 0.81169
Affinity: 31.74307 1.84532 (kcal/mol)
RMSD: 0.61906
CNNscore: 0.13388
CNNaffinity: 7.43139
CNNvariance: 0.48966
Affinity: 23.85833 0.74919 (kcal/mol)
RMSD: 1.20128
CNNscore: 0.30455
CNNaffinity: 7.38974
CNNvariance: 1.02986
Affinity: 28.49692 -1.56280 (kcal/mol)
RMSD: 1.44615
CNNscore: 0.11662
CNNaffinity: 7.23913
CNNvariance: 0.32035
Affinity: 30.89667 0.66039 (kcal/mol)
RMSD: 0.78106
CNNscore: 0.10622
CNNaffinity: 6.97652
CNNvariance: 0.20354
Affinity: 29.42368 -0.67339 (kcal/mol)
RMSD: 0.12195
CNNscore: 0.10429
CNNaffinity: 7.07745
CNNvariance: 0.48168
Affinity: 32.36478 -0.21984 (kcal/mol)
RMSD: 0.76341
CNNscore: 0.19677
CNNaffinity: 7.34227
CNNvariance: 0.28907
Affinity: 25.67328 -2.37170 (kcal/mol)
RMSD: 2.33649
CNNscore: 0.09625
CNNaffinity: 6.73323
CNNvariance: 0.52573
Affinity: 29.20913 -0.54543 (kcal/mol)
RMSD: 0.44565
CNNscore: 0.08706
CNNaffinity: 6.87676
CNNvariance: 0.27601
Affinity: 34.82842 -1.08318 (kcal/mol)
RMSD: 0.63733
CNNscore: 0.44400
CNNaffinity: 7.60950
CNNvariance: 0.58267
Affinity: 31.37675 -0.52925 (kcal/mol)
RMSD: 0.16133
CNNscore: 0.05333
CNNaffinity: 7.06941
CNNvariance: 0.43550
Affinity: 27.52372 -0.97973 (kcal/mol)
RMSD: 1.39163
CNNscore: 0.08281
CNNaffinity: 7.40354
CNNvariance: 0.48763
Affinity: 5.06220 -1.05273 (kcal/mol)
RMSD: 5.32985
CNNscore: 0.40105
CNNaffinity: 6.99272
CNNvariance: 0.38052
Affinity: 45.75579 -1.32408 (kcal/mol)
RMSD: 0.10918
CNNscore: 0.01102
CNNaffinity: 7.01824
CNNvariance: 0.90725
Affinity: 23.73584 3.60914 (kcal/mol)
RMSD: 3.31794
CNNscore: 0.06800
CNNaffinity: 7.19103
CNNvariance: 0.35771
Process Scores¶
When the output is an sdf file, scores are included as data fields.
from openbabel import pybel
import pandas as pd
scores = []
for mol in pybel.readfile('sdf','gnina_scored.sdf.gz'):
scores.append({'title': mol.title,
'CNNscore': float(mol.data['CNNscore']),
'CNNaffinity': float(mol.data['CNNaffinity']),
'Vinardo': float(mol.data['minimizedAffinity'])})
scores = pd.DataFrame(scores)
scores['label'] = scores.title.str.contains('active')
scores
| title | CNNscore | CNNaffinity | Vinardo | label | |
|---|---|---|---|---|---|
| 0 | 27626_CHEMBL135_active | 0.899126 | 7.911128 | -9.72913 | True |
| 1 | 224402_CHEMBL135236_active | 0.937766 | 7.649126 | -9.80264 | True |
| 2 | 410956_CHEMBL245378_active | 0.618515 | 7.485490 | -8.73188 | True |
| 3 | 21685_CHEMBL278233_active | 0.861939 | 7.897575 | -8.81335 | True |
| 4 | 396502_CHEMBL234638_active | 0.851940 | 7.480666 | -9.20967 | True |
| ... | ... | ... | ... | ... | ... |
| 371 | 203823_CHEMBL121879_active | 0.053331 | 7.069412 | 31.37675 | True |
| 372 | 323694_CHEMBL195466_active | 0.082807 | 7.403542 | 27.52372 | True |
| 373 | C63743815_decoy | 0.401051 | 6.992716 | 5.06220 | False |
| 374 | 323515_CHEMBL191646_active | 0.011020 | 7.018235 | 45.75579 | True |
| 375 | C39461463_decoy | 0.068003 | 7.191034 | 23.73584 | False |
376 rows × 5 columns
from sklearn.metrics import roc_auc_score, roc_curve, auc
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(dpi=150)
plt.plot([0,1],[0,1],'k--',alpha=0.5,linewidth=1)
fpr,tpr,_ = roc_curve(scores.label,-scores.Vinardo)
plt.plot(fpr,tpr,label="Vinardo (AUC = %.2f)"%auc(fpr,tpr))
fpr,tpr,_ = roc_curve(scores.label,scores.CNNaffinity)
plt.plot(fpr,tpr,label="CNNaffinity (AUC = %.2f)"%auc(fpr,tpr))
fpr,tpr,_ = roc_curve(scores.label, scores.CNNaffinity.rank() + (-scores.Vinardo).rank())
plt.plot(fpr,tpr,label="Consensus (AUC = %.2f)"%auc(fpr,tpr))
plt.legend(loc='lower right')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.gca().set_aspect('equal')
Custom Scoring with GNINA Descriptors¶
!gnina -r errec.pdb -l minimized_results.sdf.gz --score_only --custom_scoring everything.txt > scores.txt 2>&1
!head -30 scores.txt
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders
_
(_)
__ _ _ __ _ _ __ __ _
/ _` | '_ \| | '_ \ / _` |
| (_| | | | | | | | | (_| |
\__, |_| |_|_|_| |_|\__,_|
__/ |
|___/
gnina master:aab122e+ Built Apr 12 2021.
gnina is based on smina and AutoDock Vina.
Please cite appropriately.
Commandline: gnina -r errec.pdb -l minimized_results.sdf.gz --score_only --custom_scoring everything.txt
## Name gauss(o=0,_w=0.5,_c=8) gauss(o=3,_w=2,_c=8) repulsion(o=0,_c=8) hydrophobic(g=0.5,_b=1.5,_c=8) non_hydrophobic(g=0.5,_b=1.5,_c=8) vdw(i=4,_j=8,_s=0,_^=100,_c=8) vdw(i=6,_j=12,_s=1,_^=100,_c=8) non_dir_h_bond(g=-0.7,_b=0,_c=8) non_dir_anti_h_bond_quadratic(o=0.4,_c=8) non_dir_h_bond_lj(o=-0.7,_^=100,_c=8) acceptor_acceptor_quadratic(o=0,_c=8) donor_donor_quadratic(o=0,_c=8) ad4_solvation(d-sigma=3.6,_s/q=0.01097,_c=8) ad4_solvation(d-sigma=3.6,_s/q=0,_c=8) electrostatic(i=1,_^=100,_c=8) electrostatic(i=2,_^=100,_c=8) num_tors_div num_heavy_atoms_div num_heavy_atoms num_tors_add num_tors_sqr num_tors_sqrt num_hydrophobic_atoms ligand_length
Affinity: 206.20950 (kcal/mol)
CNNscore: 0.89098
CNNaffinity: 7.88183
CNNvariance: 0.70599
Intramolecular energy: 0.00000
Term values, before weighting:
## 27626_CHEMBL135_active 68.14853 1297.00342 1.76794 69.30365 32.46139 -450.29480 -509.33688 2.59811 0.00000 -18.78727 0.30538 0.47367 -12.17058 -63.88084 0.25889 0.06443 0.00000 0.00000 1.00000 0.00000 0.00000 0.00000 0.80000 0.00000
Affinity: 213.01897 (kcal/mol)
CNNscore: 0.92071
CNNaffinity: 7.73812
CNNvariance: 0.54183
import subprocess, io, re
terms = pd.read_csv(io.BytesIO(subprocess.check_output("grep \#\# scores.txt | sed 's/## //'",shell=True)),delim_whitespace=True)
terms[['CNNscore','CNNaffinity','CNNvariance']] = re.findall(r'CNNscore: (\S+)\s*CNNaffinity: (\S+)\s*CNNvariance: (\S+)',open('scores.txt').read())
terms['label'] = terms.Name.str.contains('active')
terms
| Name | gauss(o=0,_w=0.5,_c=8) | gauss(o=3,_w=2,_c=8) | repulsion(o=0,_c=8) | hydrophobic(g=0.5,_b=1.5,_c=8) | non_hydrophobic(g=0.5,_b=1.5,_c=8) | vdw(i=4,_j=8,_s=0,_^=100,_c=8) | vdw(i=6,_j=12,_s=1,_^=100,_c=8) | non_dir_h_bond(g=-0.7,_b=0,_c=8) | non_dir_anti_h_bond_quadratic(o=0.4,_c=8) | ... | num_heavy_atoms | num_tors_add | num_tors_sqr | num_tors_sqrt | num_hydrophobic_atoms | ligand_length | CNNscore | CNNaffinity | CNNvariance | label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 27626_CHEMBL135_active | 68.14853 | 1297.00342 | 1.76794 | 69.30365 | 32.46139 | -450.29480 | -509.33688 | 2.59811 | 0.00000 | ... | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.89098 | 7.88183 | 0.70599 | True |
| 1 | 224402_CHEMBL135236_active | 65.79678 | 1306.20532 | 1.14326 | 65.56741 | 31.95742 | -451.03317 | -507.68082 | 1.84834 | 0.00000 | ... | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.92071 | 7.73812 | 0.54183 | True |
| 2 | 410956_CHEMBL245378_active | 72.39946 | 1418.66785 | 3.75621 | 80.18884 | 30.10270 | -483.51788 | -550.93170 | 3.25625 | 0.00000 | ... | 1.10 | 0.0 | 0.00 | 0.00000 | 0.90 | 0.0 | 0.62149 | 7.47755 | 0.09851 | True |
| 3 | 21685_CHEMBL278233_active | 75.66557 | 1297.38098 | 1.95984 | 69.39748 | 33.11440 | -449.71274 | -504.95129 | 2.08019 | 0.63964 | ... | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.85959 | 7.98242 | 0.97652 | True |
| 4 | 396502_CHEMBL234638_active | 68.18975 | 1358.41638 | 2.36499 | 68.17257 | 35.15848 | -470.74600 | -537.57410 | 2.89272 | 0.00000 | ... | 1.05 | 1.0 | 0.02 | 0.04472 | 0.70 | 1.0 | 0.85391 | 7.61320 | 0.63155 | True |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 371 | 203823_CHEMBL121879_active | 158.93176 | 2166.97583 | 84.03927 | 97.89775 | 99.74195 | 533.27856 | -891.06244 | 2.99093 | 0.22302 | ... | 1.60 | 3.0 | 0.18 | 0.07746 | 0.70 | 4.0 | 0.05283 | 7.09754 | 0.39419 | True |
| 372 | 323694_CHEMBL195466_active | 152.78558 | 2303.83032 | 94.87981 | 127.94479 | 56.98870 | 355.76010 | -825.85254 | 2.64426 | 0.43116 | ... | 1.75 | 4.0 | 0.32 | 0.08944 | 0.95 | 6.0 | 0.08327 | 7.50873 | 0.36746 | True |
| 373 | C63743815_decoy | 168.60873 | 2075.61182 | 98.44007 | 105.16328 | 96.86362 | 591.63190 | -721.57538 | 2.55035 | 0.45564 | ... | 1.60 | 5.0 | 0.50 | 0.10000 | 0.85 | 6.0 | 0.20179 | 6.65454 | 0.61570 | False |
| 374 | 323515_CHEMBL191646_active | 197.59131 | 2406.05493 | 134.88353 | 151.78667 | 86.15619 | 943.94849 | -710.51605 | 3.06282 | 0.73817 | ... | 1.80 | 4.0 | 0.32 | 0.08944 | 1.00 | 6.0 | 0.01218 | 7.03855 | 0.83434 | True |
| 375 | C39461463_decoy | 178.48976 | 2389.42480 | 207.31912 | 125.23049 | 80.81989 | 2416.96338 | -139.25075 | 3.19046 | 2.44244 | ... | 1.85 | 8.0 | 1.28 | 0.12649 | 1.00 | 10.0 | 0.03426 | 6.07510 | 1.61250 | False |
376 rows × 29 columns
import sklearn
from sklearn.linear_model import *
X = terms.drop(['Name','label'],axis=1).astype(float) # features
Y = terms.label
X
| gauss(o=0,_w=0.5,_c=8) | gauss(o=3,_w=2,_c=8) | repulsion(o=0,_c=8) | hydrophobic(g=0.5,_b=1.5,_c=8) | non_hydrophobic(g=0.5,_b=1.5,_c=8) | vdw(i=4,_j=8,_s=0,_^=100,_c=8) | vdw(i=6,_j=12,_s=1,_^=100,_c=8) | non_dir_h_bond(g=-0.7,_b=0,_c=8) | non_dir_anti_h_bond_quadratic(o=0.4,_c=8) | non_dir_h_bond_lj(o=-0.7,_^=100,_c=8) | ... | num_heavy_atoms_div | num_heavy_atoms | num_tors_add | num_tors_sqr | num_tors_sqrt | num_hydrophobic_atoms | ligand_length | CNNscore | CNNaffinity | CNNvariance | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 68.14853 | 1297.00342 | 1.76794 | 69.30365 | 32.46139 | -450.29480 | -509.33688 | 2.59811 | 0.00000 | -18.78727 | ... | 0.0 | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.89098 | 7.88183 | 0.70599 |
| 1 | 65.79678 | 1306.20532 | 1.14326 | 65.56741 | 31.95742 | -451.03317 | -507.68082 | 1.84834 | 0.00000 | -14.06621 | ... | 0.0 | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.92071 | 7.73812 | 0.54183 |
| 2 | 72.39946 | 1418.66785 | 3.75621 | 80.18884 | 30.10270 | -483.51788 | -550.93170 | 3.25625 | 0.00000 | -19.31251 | ... | 0.0 | 1.10 | 0.0 | 0.00 | 0.00000 | 0.90 | 0.0 | 0.62149 | 7.47755 | 0.09851 |
| 3 | 75.66557 | 1297.38098 | 1.95984 | 69.39748 | 33.11440 | -449.71274 | -504.95129 | 2.08019 | 0.63964 | -15.31258 | ... | 0.0 | 1.00 | 0.0 | 0.00 | 0.00000 | 0.80 | 0.0 | 0.85959 | 7.98242 | 0.97652 |
| 4 | 68.18975 | 1358.41638 | 2.36499 | 68.17257 | 35.15848 | -470.74600 | -537.57410 | 2.89272 | 0.00000 | -20.88869 | ... | 0.0 | 1.05 | 1.0 | 0.02 | 0.04472 | 0.70 | 1.0 | 0.85391 | 7.61320 | 0.63155 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 371 | 158.93176 | 2166.97583 | 84.03927 | 97.89775 | 99.74195 | 533.27856 | -891.06244 | 2.99093 | 0.22302 | 23.77504 | ... | 0.0 | 1.60 | 3.0 | 0.18 | 0.07746 | 0.70 | 4.0 | 0.05283 | 7.09754 | 0.39419 |
| 372 | 152.78558 | 2303.83032 | 94.87981 | 127.94479 | 56.98870 | 355.76010 | -825.85254 | 2.64426 | 0.43116 | 15.61960 | ... | 0.0 | 1.75 | 4.0 | 0.32 | 0.08944 | 0.95 | 6.0 | 0.08327 | 7.50873 | 0.36746 |
| 373 | 168.60873 | 2075.61182 | 98.44007 | 105.16328 | 96.86362 | 591.63190 | -721.57538 | 2.55035 | 0.45564 | 1.18331 | ... | 0.0 | 1.60 | 5.0 | 0.50 | 0.10000 | 0.85 | 6.0 | 0.20179 | 6.65454 | 0.61570 |
| 374 | 197.59131 | 2406.05493 | 134.88353 | 151.78667 | 86.15619 | 943.94849 | -710.51605 | 3.06282 | 0.73817 | -20.87544 | ... | 0.0 | 1.80 | 4.0 | 0.32 | 0.08944 | 1.00 | 6.0 | 0.01218 | 7.03855 | 0.83434 |
| 375 | 178.48976 | 2389.42480 | 207.31912 | 125.23049 | 80.81989 | 2416.96338 | -139.25075 | 3.19046 | 2.44244 | 190.20380 | ... | 0.0 | 1.85 | 8.0 | 1.28 | 0.12649 | 1.00 | 10.0 | 0.03426 | 6.07510 | 1.61250 |
376 rows × 27 columns
model = LogisticRegression(solver='liblinear')
cvpredict = sklearn.model_selection.cross_val_predict(model, X, Y, method='predict_proba')
fpr,tpr,_ = roc_curve(Y,cvpredict[:,1])
fig,ax = plt.subplots(1,1,dpi=100)
ax.plot(fpr,tpr,label="Combined CV ROC (AUC=%.2f)"%auc(fpr,tpr))
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.legend()
ax.set_aspect('equal');
%%html
<div id="mpredict" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#mpredict';
jQuery(divid).asker({
id: divid,
question: "Would this model be useful for further screening?",
answers: ['Yes','No','Eh'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".input .o:contains(html)").closest('.input').hide();
</script>
Skepticism is always warranted with learned models¶

- Should rigorously cross-validate (e.g. scaffold split)
- Need training sets that accurately represent screening library
- Actives and decoys should not be trivially seperable
- Structure-based models should be pose-sensitive
- Interrogate model (easy with empirical features)
Do not use virtual screening classification models to dock/minimize!¶
Training CNN Models¶
Beyond the scope of this workshop (sorry!)
Scripts and documentation here: https://github.com/gnina/scripts
Need to get data into a text file with this format:
<label> <affinity> <receptor> <ligand>Then design and train models in Caffe.